1727
A generalized deep learning network for fractional anisotropy reconstruction: application to epilepsy and multiple sclerosis1Department of Electrical, Computer and Biomedical Engineering, University of Pavia, Pavia, Italy, 2NMR Research Unit, Queen Square Multiple Sclerosis Centre, Department of Neuroinflammation, UCL Queen Square Institute of Neurology, UCL, London, United Kingdom, 3Department of Brain and Behavioural Sciences, University of Pavia, Pavia, Italy, 4IRCCS Policlinico San Donato, Milano, Italy, 5Brain Connectivity Centre, IRCCS Mondino Foundation, Pavia, Italy
Synopsis
Quantitative maps obtained from Diffusion Tensor are very useful for investigating microstructural changes that occur in brain diseases. However, the long acquisition times required for a fully sampled diffusion-weighted space makes their clinical use unfeasible. Here we have adapted a U-net that obtains reliable Fractional Anisotropy (FA) maps from a reduced set of 10 Diffusion Weighted volumes (that can be acquired in less than 1 min). Our network was applied to two independent, clinical datasets, without retraining, and produced FA that retained clinical sensitivity and characteristic FA value distributions in the brain white matter.
INTRODUCTION
Diffusion Tensor Imaging is a Magnetic Resonance Imaging method sensitive to brain microstructure and can be used clinically to investigate pathological alterations. Theoretically, 7 diffusion-weighted (DW) images are required to estimate the diffusion tensor (DT) with model fitting, but in practice it has been widely demonstrated that increasing the number of DW volumes (e.g. to 35 or 601,2) results in less noisy and biased DT estimates. This inevitably leads to long acquisition times limiting the DT clinical viability. The most used quantitative map obtained from the DT is fractional anisotropy (FA), which is highly sensitive to changes in tissue microstructure3,4.Here we investigate the possibility of reducing the number of DW volumes to dramatically reduce acquisition time, through deep learning (DL), while retaining or enhancing image quality exploiting the fact that a DL network can contribute to image quality transfer5. We implemented a DL network that can: 1) map from a small number of DW volumes (10) to FA; 2) take advantage of datasets with high-geometrical and DW angular resolution with corresponding high-quality FA maps for training the network, e.g using data from the Human Connectome Project (HCP)6; 3) tackle the generalization problem: given a network trained with a specific dataset, offer applicability to clinical datasets from different scanners without re-training; and, 4) retain sensitivity to pathology. We validated the network with unseen data from the HCP dataset and with two datasets from temporal lobe epilepsy (TLE) and multiple sclerosis (MS) studies, acquired with different protocols on different scanners.
METHODS
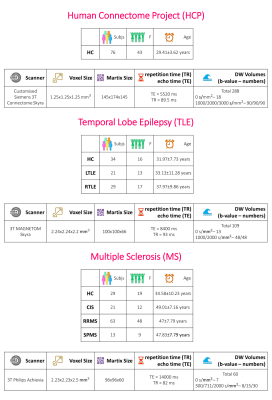
Datasets: Fig.1 shows the demographics and MRI acquisition details. A standard FA map, from DT fitting (DESIGNER7), was calculated considering all DW volumes for each dataset.Data preparation: The HCP dataset included 54 subjects for training, 11 for validation and 11 for test. Standard FA maps were used as ground-truth (GT). We cropped the images to 128x160 voxels. We normalized the DWI intensity for each subject separately, considering all volumes together.
We used Camino8 to select subsets of b-vectors equally distributed in space, obtaining 10 different DW-subsets containing 9 volumes with b-value=1000s/mm2 to which one random volume with b-value=0 was added. We also calculated the standard FA from one subset.
Network architecture: We adapted the U-net architecture9: the input was one of the 10 DW subsets, where each volume was a channel, with a single output channel to FA. The loss function was the root mean square error (RMSE).
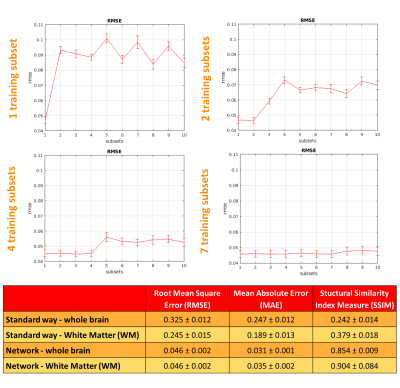
Training: The best combination of hyperparameters was the one with the lowest mean RMSE on the validation subjects. The choice of DW inputs is crucial for estimating FA: we trained the network 4 times by increasing the number of DW subsets as input (1,2,4,7) each matching the same FA (GT).
Performance assessment: RMSE, mean absolute error and structural similarity index measure10 were calculated at voxel level between the GT and the network output considering only brain voxels. To assess generalisability to unseen DW volumes combinations, each of the 4 trained networks were applied to each of the 10 DW-subsets of the test subjects. We chose the network with the most similar performance, e.g. similar RMSE, using the DW training subsets and the remaining DW subsets.
Clinical Sensitivity: Our ultimate aim was to use our network with clinical datasets, acquired with limited DW volumes and coarser resolution compared to HCP data, to maintain or improve FA quality and sensitivity to pathological changes. We used the TLE and MS datasets to evaluate network performance. Per dataset, DW data were resampled and cropped to HCP resolution, the 9 most non-collinear directions were chosen (Camino) and our network was applied.
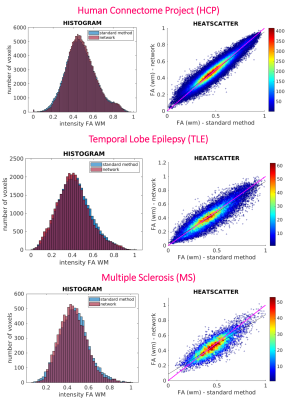
White matter (WM) quantitative assessment: WM-FA values from standard-full-DW and network-subset-DW data were compared at subject-level using histogram and heatscatter plots and at group-level using boxplot of subject’s mean WM-FA. For clinical datasets, statistics between groups verified whether biases were introduced in WM-FA when using the network-FA compared to GT.
RESULTS
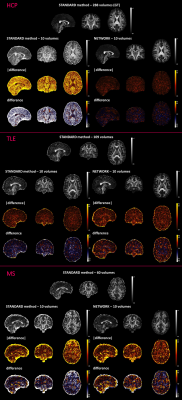
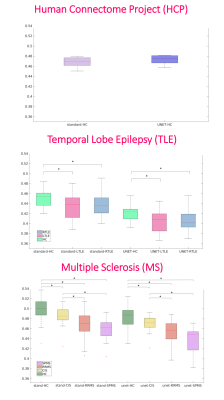
We selected the network trained with 7 DW subsets and we reported its performance metrics for the HCP test subjects (Fig.2). Fig.3 shows for each dataset each FA (standard-full-DW, standard-subset-DW, network-subset-DW). Fig.4 shows WM-FA plots in a single-subject and Fig.5 the boxplots at group-level with group differences indicated when significant.DISCUSSION & CONCLUSIONS
We implemented a network capable of obtaining FA from a reduced set of 10 DW volumes. For HCP, the network-FA (RMSE = 0.046) was much closer to the GT than the standard-FA (DT fitting) from the same 10 DW volumes (RMSE = 0.325).Our network maintained FA properties even on clinical data acquired on different scanners, with different DW directions and different b-values. Single-subjects FA distribution in WM voxels was maintained when using the network FA for all datasets (Fig.4).
The network FA retained properties of the standard-FA with all volumes, including sensitivity to pathology. Significant pathological differences between groups found when using the full DW dataset for standard-FA remained significant when using FA estimated by the network.
Acquiring only 10 DWI greatly reduces acquisition time and reconstruction with our network could enable the use of FA in the clinic. Future works will aim to reconstruct other DT maps of tissue microstructure in addition to FA.
Acknowledgements
This research received funding by H2020 Research and Innovation Action Grants Human Brain Project 785907 and 945539 (SGA2 and SGA3) and by the MNL Project “Local Neuronal Microcircuits” of the Centro Fermi (Rome, Italy) to FP. CGWK received funding from the UK MS Society (#77), Wings for Life (#169111), Horizon2020 (CDS-QUAMRI, #634541), BRC (#BRC704/CAP/CGW).References
1. Zhan, L. et al. HOW DOES ANGULAR RESOLUTION AFFECT DIFFUSION. 49, 1357–1371 (2011).
2. Landman, B. A. et al. Effects of diffusion weighting schemes on the reproducibility of DTI-derived fractional anisotropy, mean diffusivity, and principal eigenvector measurements at 1.5T. Neuroimage 36, 1123–1138 (2007).
3. Alexander, A. L., Lee, J. E., Lazar, M. & Field, A. S. Diffusion Tensor Imaging of the Brain. Neurotherapeutics 4, 316–329 (2007).
4. Giannelli, M. et al. Dependence of brain DTI maps of fractional anisotropy and mean diffusivity on the number of diffusion weighting directions. J. Appl. Clin. Med. Phys. 11, 176–190 (2010).
5. Alexander, D. C. et al. Image quality transfer and applications in diffusion MRI. Neuroimage 152, 283–298 (2017).
6. Van Essen, D. C. et al. The WU-Minn Human Connectome Project: An overview. Neuroimage 80, 62–79 (2013).
7. Ades-Aron, B. et al. Evaluation of the accuracy and precision of the diffusion parameter EStImation with Gibbs and NoisE removal pipeline. Neuroimage 183, 532–543 (2018).
8. Cook, P. A. et al. Camino: Diffusion MRI reconstruction and processing. Statistics (Ber). (2005).
9. Ronneberger, O., Fischer, P. & Brox, T. U-Net: Convolutional Networks for Biomedical Image Segmentation. in Medical Image Computing and Computer-Assisted Intervention -- MICCAI 2015 (eds. Navab, N., Hornegger, J., Wells, W. M. & Frangi, A. F.) 234–241 (Springer International Publishing, 2015).
10. Wang, Z., Bovik, A. C., Sheikh, H. R. & Simoncelli, E. P. Image quality assessment: From error visibility to structural similarity. IEEE Trans. Image Process. 13, 600–612 (2004).
Figures