1726
Three-component IVIM fitting in cerebrovascular disease using physics-informed neural networks: repeatability and accuracy1Department of Radiology & Nuclear Medicine, Maastricht University Medical Center, Maastricht, Netherlands, 2School for Mental Health & Neuroscience, Maastricht University, Maastricht, Netherlands, 3School for Cardiovascular Disease, Maastricht University, Maastricht, Netherlands, 4Department of Radiology and Nuclear medicine, Amsterdam University Medical Center, Cancer Center Amsterdam, University of Amsterdam, Amsterdam, Netherlands, 5Department of Neurology, Maastricht University Medical Center, Maastricht, Netherlands, 6Department of Electrical Engineering, Eindhoven University of Technology, Eindhoven, Netherlands
Synopsis
Next to parenchymal diffusion and microvascular pseudo-diffusion, a third diffusion component is present in cerebral intravoxel incoherent motion (IVIM) imaging, representing interstitial fluid. Fitting the three-component IVIM model using conventional fitting methods strongly suffers from image noise. Therefore, we explored the applicability of a physics-informed neural network (PI-NN) fitting approach, previously shown to be more robust to noise. Using test-retest data from sixteen patients with cerebrovascular disease, we found higher repeatability of all IVIM parameters using PI-NN. Furthermore, simulations showed that PI-NN provided more accurate IVIM parameters. Hence, using PI-NN is promising to obtain tissue markers of cerebrovascular disease.
Introduction
Intravoxel incoherent motion (IVIM) imaging is a diffusion-weighted MR technique, which can simultaneously measure the parenchymal diffusion (Dpar) and microvascular pseudo-diffusion (Dmv).1 Recently, the presence of a third, intermediate diffusion component (Dint) was revealed, which was suggested to represent interstitial fluid (ISF) within the parenchyma.2,3 Dint was furthermore found to be related to cerebral small vessel disease (cSVD) markers, such as white matter hyperintensities (WMH).2 However, with current fitting techniques, extracting accurate voxel-wise measures for the three diffusion components is extremely hard due to the strong influence of noise. Recently, Troelstra et al.4 showed that fitting a tri-exponential IVIM model using an unsupervised physics-informed neural network (PI-NN) resulted in more robust voxel-wise IVIM parameters compared to the conventional least squares (LSQ) method in the liver.In this study, we have adapted the three-component PI-NN to estimate the cerebral diffusion components (Dpar, Dint and Dmv). We aimed to assess the performance of our PI-NN compared to conventional LSQ in terms of test-retest repeatability (using in-vivo data of patients with cerebrovascular disease) and accuracy (using simulations).
Methods
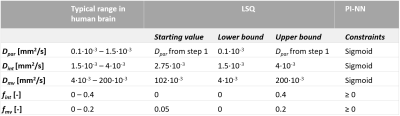
In-vivo data: Sixteen patients with cerebrovascular disease (cSVD (n=11) and cortical stroke (n=5)) underwent brain MRI twice on separate days (Philips Achieva 3.0T). IVIM images were acquired with diffusion sensitization in three orthogonal directions, and included an inversion pulse for cerebrospinal fluid suppression (single-shot spin-echo echo-planar-imaging (EPI), 2.4 mm cubic voxel size, b-values: 0,5,7,10,15,20,30,40,50,60,100,200,400,700, and 1000 s/mm2). Additionally, T2-weighted FLAIR and T1-weighted images were acquired for anatomical reference.Image analysis: The IVIM images were corrected for head displacement, EPI and eddy current distortions (ExploreDTI). The three-component cerebral IVIM model, accounting for the inversion recovery, was described as:2
\begin{equation}S(b)=S(0)*[\frac{(1-f_{mv}-f_{int})E_{1,par}E_{2,par}e^{-bD_{par}}+f_{int}E_{1,int}E_{2,int}e^{-bD_{int}}+f_{mv}E_{1,mv}E_{2,mv}e^{-bD_{mv}}}{(1-f_{mv}-f_{int})E_{1,par}E_{2,par}+f_{int}E_{1,int}E_{2,int}+f_{mv}E_{1,mv}E_{2,mv}}\;\;\;\;\;\;\;\;\;[Eq.\;1]\end{equation}\begin{equation}with\;E_{1,k}=(1-2e^{-\frac{TI}{T_{1,k}}}+e^{-\frac{TR}{T_{1,k}}})\;for\;k=par,int;\;E_{1,mv}=(1-e^{-\frac{TR}{T_{1,blood}}});\;E_{2,k}=(e^{-\frac{TE}{T_{2,k}}})\;for\;k=par,int,mv)\end{equation}
Here, fmv and fint are the volume fractions of the Dmv and Dint components, respectively. Both LSQ and PI-NN fitting approaches were applied to the Trace images in a voxel-wise manner. For the LSQ approach, a two-step Levenberg-Marquardt algorithm was used with lower and upper bounds equal to typical ranges found with IVIM in the human brain (Table 1).2 For the PI-NN approach, the hyperparameters and training features were based on a previous study with some small adjustments to make it suitable for brain data.5 For example, the loss function was changed according to Equation 1. The diffusivity parameters were constrained by a sigmoid function, and the fractions were constrained to be positive (Table 1). To mitigate the problem of variable results due to repeated training, we trained 20 instances of the PI-NN model on all brain voxels in the in-vivo dataset and averaged the corresponding predictions.5 The white matter (WM) and cortical gray matter (cGM) were automatically segmented from the T1-weighted images using Freesurfer, whereas WMH were semi-automatically segmented from the FLAIR images.6
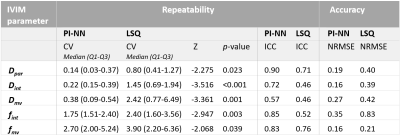
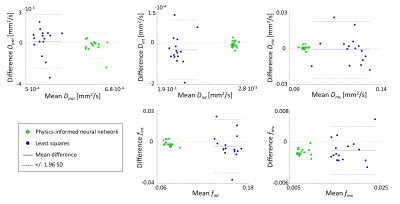
Statistical analysis: After voxel-wise fitting, the IVIM parameters were averaged over all brain voxels per subject scan. As a measure of precision, the test-retest repeatability of these IVIM parameters was quantified using the within-subject coefficient of variation (CV) and the intraclass correlation coefficient (ICC) (calculated using a one-way random model).7 Statistical difference between the CVs obtained with LSQ and PINN was tested with a paired Wilcoxon signed-rank test. Furthermore, Bland-Altman plots were created to visually compare the effect of PI-NN and LSQ on the repeatability.
Simulated data: 10.000 IVIM signal curves were generated using Equation 1, while randomly sampling the five IVIM parameters from a Gaussian distribution (97.7% within the ranges given in Table 1). Gaussian noise was added, similar to the noise level in our in-vivo data (signal-to-noise ratio at b=0 s/mm2 was 35).8 An ensemble of 20 PI-NN models was trained on 1.000.000 noisy IVIM signal decay curves. Subsequently, the PI-NN and LSQ fitting approaches were applied to the synthesized curves to predict the IVIM parameters. The accuracy of both methods was quantified with the normalized root-mean-square error (NRMSE) between all ground-truth and predicted IVIM parameters.
Results
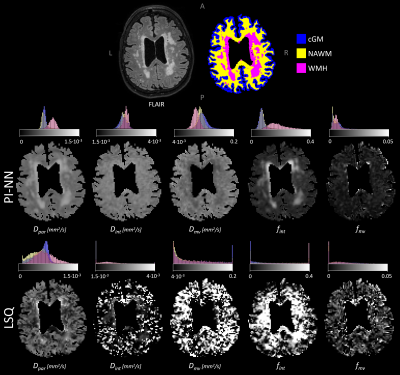
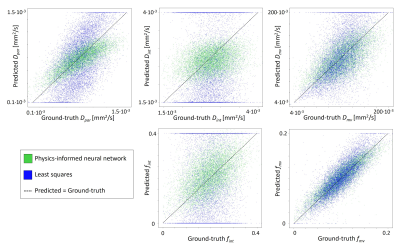
In-vivo data: The IVIM parameter maps obtained with the PI-NN and LSQ fitting approach of a representative cSVD patient are shown in Figure 1. The PI-NN maps appear smoother, while still retaining reasonable contrast between different tissue types (e.g. higher Dpar, higher fint and lower Dmv in WMH compared to normal-appearing WM). The PI-NN method was more precise than the LSQ, as it showed higher test-retest repeatability in terms of lower CV and higher ICC values (Table 2). This was also illustrated by the smaller differences between the test-retest of the PI-NN measurements using Bland-Altman plots (Figure 2).Simulated data: Figure 3 shows scatterplots of the ground-truth and predicted IVIM parameters. Overall, the PI-NN was more accurate as it had a lower NRMSE than the LSQ (Table 2).
Discussion & Conclusion
In this study, we demonstrated the applicability of a PI-NN for three-component cerebral IVIM fitting in patients with cerebrovascular disease. The PI-NN method resulted in more precise (repeatable) and more accurate IVIM parameters as compared to the LSQ method. The high-quality parameter maps generated with PI-NNs are needed for visual evaluation of increased ISF (Dint, fint), microstructural damage (Dpar) and microvascular alterations (Dmv, fmv) within patients, and enables analysis of these parameters on a voxel-wise basis, allowing assessment of small WMH lesions or perilesional WM.Acknowledgements
This work has received funding from the European Union’s Horizon 2020 research and innovation programme ‘CRUCIAL’ under grant number 848109.References
1. Le Bihan D, Breton E, Lallemand D, Grenier P, Cabanis E, Laval-Jeantet M. MR imaging of intravoxel incoherent motions: application to diffusion and perfusion in neurologic disorders. Radiology. 1986;161(2):401-7.
2. Wong SM, Backes WH, Drenthen GS, Zhang CE, Voorter PH, Staals J, et al. Spectral diffusion analysis of intravoxel incoherent motion MRI in cerebral small vessel disease. Journal of Magnetic Resonance Imaging. 2020;51(4):1170-80.
3. van der Thiel MM, Freeze WM, Verheggen IC, Wong SM, de Jong JJ, Postma AA, et al. Associations of increased interstitial fluid with vascular and neurodegenerative abnormalities in a memory clinic sample. Neurobiology of Aging. 2021;106:257-67
4. Troelstra M, Witjes J, van Dijk AM, Mak AL, Runge J, Verheij J, et al. Physics-informed deep neural network for tri-exponential intravoxel incoherent motion fitting in non-alcoholic fatty liver disease. Proceedings of the 30th annual meeting of the ISMRM 2021:0312
5. Kaandorp MP, Barbieri S, Klaassen R, van Laarhoven HW, Crezee H, While PT, et al. Improved unsupervised physics‐informed deep learning for intravoxel incoherent motion modeling and evaluation in pancreatic cancer patients. Magnetic Resonance in Medicine. 2021;86(4):2250-65.
6. De Boer R, Vrooman HA, Van Der Lijn F, Vernooij MW, Ikram MA, Van Der Lugt A, et al. White matter lesion extension to automatic brain tissue segmentation on MRI. Neuroimage. 2009;45(4):1151-61.
7. Liljequist D, Elfving B, Skavberg Roaldsen K. Intraclass correlation–A discussion and demonstration of basic features. PloS one. 2019;14(7):e0219854.
8. National Electrical Manufacturers Association. Determination of signal-to-noise ratio (SNR) in diagnostic magnetic resonance imaging. NEMA Standards Publication MS 1-2008. 2008.
Figures