1691
Comparison of New MR Approaches for Accelerated Knee Imaging1Radiology, Stanford University, Stanford, CA, United States, 2GE Healthcare, New York, NY, United States, 3GE Healthcare, Milwaukee, WI, United States
Synopsis
Conventional knee MRI protocols use numerous 2D-FSE sequences with multiple contrasts, which have long scan times and don’t fully use modern parallel imaging and improved SNR technologies.In this abstract, we outline four approaches that reduce protocol times to <6 minutes and potentially add diagnostic value through quantitative data or oblique reformats.These include an accelerated conventional 2D protocol, SNR-efficient 3D qDESS and CUBE approaches, and a thin-Slice protocol with high slice resolution.We propose a study that will evaluate the performance of each of these knee MRI protocols, and determine diagnostic utility and confidence with respect to fluid sensitivity and tissue pathologies.
Introduction
MRI is the hallmark for diagnostic knee imaging due to its excellent soft-tissue sensitivity with multiple contrasts. Conventional knee MRI protocols use numerous 2D fast spin echo (FSE) sequences with multiple contrasts, often repeated in multiple scan planes. This inefficient approach requires longer scan times and does not fully utilize modern 3D, parallel imaging and improved coil SNR technologies. Numerous approaches have been proposed to improve the value of knee imaging by reducing protocol times to less than 6 minutes and perhaps adding diagnostic potential through quantitative data or oblique reformats. In this abstract, we outline four such approaches: an accelerated conventional 2D protocol; SNR-efficient 3D qDESS and CUBE protocols; and a thin-Slice protocol with high slice resolution. The proposed study will aim to evaluate the performance of each of these four knee MRI protocols, and determine diagnostic utility and confidence with respect to fluid sensitivity, and ligament, meniscus and cartilage pathology.Conventional (Accelerated) Knee Protocol
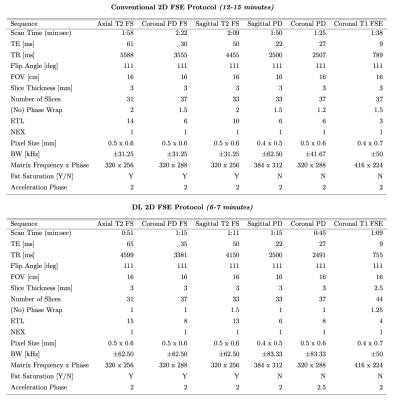
This reference 12-15 minute protocol includes six accelerated sequences[1], including axial T2-weighted fat-saturated (FS), coronal proton density (PD)-weighted FS, sagittal T2-weighted FS, sagittal PD, coronal PD, and coronal T1-weighted FSE images(parameters in Fig-1). This protocol is representative of a conventional clinical knee protocol, although these sequences may be accelerated compared to those often utilized and the number of sequences used may vary. This protocol serves as the reference standard for the study.New Approach 1: DL 2D FSE Protocol
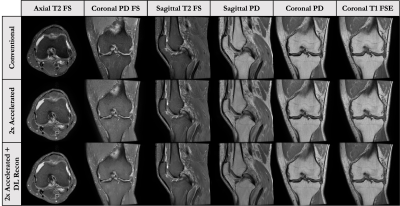
This 6-7 minute protocol includes six further accelerated sequences that were reconstructed using the GE AIR deep learning (DL) reconstruction. It includes the same sequences as the conventional 2D FSE protocol, with modified parallel imaging acceleration, bandwidth, ETL and no-phase-wrap factors to reduce scan times by roughly a factor of 2(parameters in Fig-1). Figure 3 shows representative images of the acquired and DL-reconstructed images compared to images acquired with the conventional protocol. This approach offers familiar contrasts and sequences to the current conventional protocol, and has shown promise for diagnostic equivalence[2,3].Approach 2: 3D qDESS Protocol
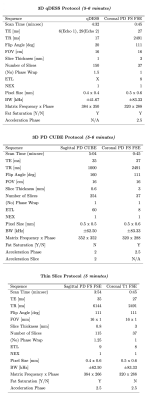
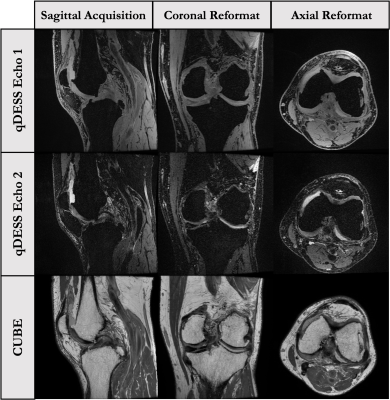
This 5-6 minute approach is based around a 3D quantitative Double-Echo Steady-State (qDESS) sequence(parameters in Fig-2, images in Fig-5). The qDESS approach offers multiple contrasts similar to the PD and T2-weighted signals with fat-saturation in conventional protocols[4]. This approach has shown near diagnostic equivalence to the conventional protocol, particularly when paired with a 2D coronal PD-FS-FSE sequence for evaluation of bone marrow lesions (BMLs) and collateral ligaments(this study uses the <1min accelerated coronal PD-FS-FSE sequence). The high SNR efficiency allows higher through-plane resolution which may support numerous oblique reformats. DL-based super resolution methods could further improve resolution to <0.5mm isotropic voxels. Additionally, qDESS also provides synovitis hybrid images (joint-fluid-suppressed images created from qDESS images) and cartilage T2 relaxation time maps, which may be beneficial for improved evaluation of synovitis and early cartilage degeneration, respectively[5].Approach 3: 3D PD CUBE Protocol
This 5-6 minute protocol includes a 3D PD-FSE sequence with variable flip-angle refocusing (CUBE) and a 2D Cor PD-FS-FSE sequence for visualization of BMLs(parameters in Fig-2, images in Fig-4). Like 3D qDESS, the 3D CUBE sequence has higher SNR-efficiency, allowing near isotropic acquisitions that provide the ability to be reformatted in any plane[6]. Further, 3D CUBE offers familiar FSE contrast to that of the conventional protocol. However, the long echo train lengths (ETLs) used in 3D-FSE sequences lead to some blurring which has limited clinical adoption in the past. New optimized parameters, including MSK-specific refocusing pulse-trains, offer improvement over previous approaches, particularly when no fat saturation is used, allowing shorter ETLs within normal scan times. The addition of a fast 2D Cor PD-FS-FSE offers fat-suppressed images for evaluation of BMLs and Hoffa’s fat pad synovitis.Approach 4: 3D Thin-Slice Protocol
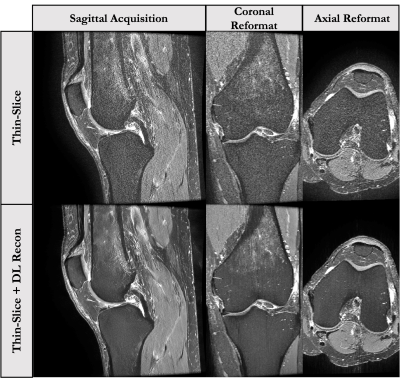
This 5-minute protocol includes a 2D sagittal PD-FS-FSE sequence with a 4x improvement in slice resolution[7] and a 2D Cor T1-FSE sequence(parameters in Fig-2). The thin-slice 2D FS-FSE sequence offers familiar contrast in a near isotropic resolution similar to that of 3D-FSE approaches without concerns of blurring due to long ETLs. However, the increased slice resolution comes at a large SNR cost. To compensate for this, the GE DL-Based recon is applied. Figure 5 shows the original and DL-Recon images acquired sagittally with this approach. One concern with this approach is that the effects of DL-Recon denoising are largely in-plane, which may affect image reformats.Study Design
The comparison study will be conducted in twenty-five individuals with knee pain and degenerative osteoarthritis, scanned on a whole-body 3T MRI system (GE Healthcare) using a 16-channel receive-only phased-array FLEX coil.All participants will undergo the 5 protocols listed above in one scan session.Three radiologists will independently evaluate the retrospectively-constructed knee MRI protocols for diagnostic confidence with respect to fluid sensitivity, tissue pathology, and image artifacts for each approach.Beyond diagnostic accuracy and confidence, we hope to evaluate the utility of oblique image reformats in 3D and Thin-Slice datasets as well as the utility of new data such as synovitis hybrid images and Cartilage T2 relaxation time maps for clinical evaluation.Each radiologist will be blinded to patients and scan protocol, and will receive 5 compilations of 25 datasets with a two-week washout period in-between.Summary
This study will evaluate the performance of five proposed knee MRI protocols for providing diagnostic confidence and utility in terms of tissue pathology, quantitative data, and oblique image reformats.Acknowledgements
This work was supported by research funding from GE Healthcare.References
[1] Chaudhari AS, Kogan F, Pedoia V, Majumdar S, Gold GE, Hargreaves BA.Rapid knee MRI acquisition and analysis techniques for imaging osteoarthritis. J Magn Reson Imaging 2020; 52:1321–1339
[2] Recht MP, Zbontar J, Sodickson DK, et al. Using Deep Learning to Accelerate Knee MRI at 3 T: Results of an Interchangeability Study. AJR Am J Roentgenol. 2020;215(6):1421-1429. doi:10.2214/AJR.20.23313
[3] Del Grande F, Rashidi A, Luna R, Delcogliano M, et al. “Five-Minute Five-Sequence Knee MRI Using Combined Simultaneous Multislice and Parallel Imaging Acceleration: Comparison with 10-Minute Parallel Imaging Knee MRI.” Radiology. Radiological Society of North America vol. 299,3 (2021): 635-646. doi:10.1148/radiol.2021203655
[4] Chaudhari AS, Grissom MJ, Fang Z, et al. Diagnostic Accuracy of Quantitative Multicontrast 5-Minute Knee MRI Using Prospective Artificial Intelligence Image Quality Enhancement. AJR Am J Roentgenol. 2021;216(6):1614-1625. doi:10.2214/AJR.20.24172
[5] Thoenen J, Stevens KJ, Turmezei TD, et al. Non-contrast MRI of synovitis in the knee using quantitative DESS [published online ahead of print, 2021 May 15]. Eur Radiol. 2021;10.1007/s00330-021-08025-2. doi:10.1007/s00330-021-08025-2
[6] Li CQ, Chen W, Rosenberg JK, et al. Optimizing isotropic three-dimensional fast spin-echo methods for imaging the knee. J Magn Reson Imaging. 2014;39(6):1417-1425. doi:10.1002/jmri.24315
[7] Tokuda O, Harada Y, Shiraishi G, et al. MRI of the anatomical structures of the knee: the proton density-weighted fast spin-echo sequence vs the proton density-weighted fast-recovery fast spin-echo sequence. Br J Radiol. 2012;85(1017):e686-e693. doi:10.1259/bjr/99570113
Figures