1647
SLAM reconstruction of 31P Cardiac MRS Improves PCr/ATP Repeatability Without Requiring Cardiac Gating1OCMR, University of Oxford, Oxford, United Kingdom, 2Department of Physiology, Anatomy & Genetics, University of Oxford, Oxford, United Kingdom, 3Department of Physics, University of Oxford, Oxford, United Kingdom, 4The MR Research Centre, Aarhus University, Aarhus, Denmark
Synopsis
Using the SLAM algorithm to reconstruct non-gated acquisition weighted 31P cardiac CSI data-sets improved repeatability by 35% and reduced variation by 23%, compared to a Fourier based reconstruction of the same data-set, without requiring a change to acquisition protocol.
Introduction
Cardiac phosphorus (31P) MRS is a powerful technique for assessing metabolic derangements, as well as novel therapies for patients with heart failure in-vivo, particularly through measurement of the phosphocreatine-to-adenosine-triphosphate (PCr/ATP) ratio[1]. Unfortunately, measurements of the PCr/ATP ratio suffer from significant variability, leaving clinical studies with low statistical power, making it difficult to draw conclusions without large patient cohorts.Since the introduction of 31P MRS, numerous strategies have been pursued to improve the repeatability of the measured PCr/ATP ratio, including improving the quality of the spectral fit by increasing SNR[2,3], using cardiac/respiratory[4] gating, or improving localization accuracy[3,5].
Unfortunately, while cardiac gating has been shown to improve spectral quality[4], scan time may be increased by up to 2x if the TR and R-R interval are similar. In this work we investigate if using the SLAM reconstruction algorithm (which incorporates a segmented localizer into the reconstruction, producing one spectra per segmented region, and expanding the VOI to the whole heart), as opposed to a Fourier based method, can improve repeatability to a similar extent as cardiac gating, when reconstructing un-gated, acquisition-weighted (AW) data.
Furthermore we use the SLAM algorithm to reconstruct 31P cardiac scans of five patients with heart failure, to investigate whether the potential improved repeatability may help recognize differences in PCr/ATP in a small patient population.
Methods
Six healthy volunteers (2F/4M) were scanned twice, using a Siemens TIM Trio 3T MRI scanner (Erlangen, Germany), with a short break outside of the scan room between scans. Volunteers were positioned in the prone position, with their hearts over the coil, consisting of a 1H/31P dual tuned 26×28 cm transmit/receive loop and a dual 12×15 cm butterfly 31P receive pair[6]. CSI-matched 1H images were acquired to provide the localizer for the SLAM reconstruction, followed by two 31P acquisitions (with/without cardiac gating), consisting of a UTE-CSI sequence[7]. Three saturation bands were used to null the signal from the chest wall and liver. All 31P acquisitions were acquisition weighted, had a FOV of 240x240×200 mm, with a matrix size of 16x8x8, and in the absence of cardiac gating, took 10.5 minutes to complete with a nominal TR of 0.9 s. Cardiac gating was prospective and triggered the acquisition to coincide with diastole.The same acquisition (no cardiac gating) was used for the patient cohort (2F/3M, BMI = 36 ± 5 kg/m2, age = 69 ± 9 years) who all had heart failure with preserved ejection fraction (HFpEF).
The SLAM reconstructions were performed using an implementation of the algorithm[3] produced in MATLAB, and the CSI reconstructions were performed using a Fourier based method, spectra produced with both methods were then fitted with the OXSA Matlab toolbox AMARES implementation. For each fitted spectrum, PCr/ATP ratios and CLRBs, as well as PCr SNR and coefficients of repeatability (CoR) and variation (CoV) (for the PCr/ATP ratio) were computed. Sign-rank Wilcoxon tests were used to compare the methods with the un-gated CSI method.
Results and Discussion
An example segmentation mask is shown in Figure 1, along with the resultant SLAM reconstruction spectrum of the cardiac compartment. Each of the expected resonances is clearly visible and the SNR is sufficient to produce a high quality fit, with no visible PCr contamination from the chest wall.A table detailing all computed results is shown in Figure 2 with box-plots of the volunteer study and clinical data-set shown in Figures 3 and 4.
The gated CSI method showed a reduction in both coefficients of variation and repeatability over the un-gated CSI method in healthy volunteers, despite there being no change to the CRLB of the fit. This suggests that cardiac motion may be a significant source of variation. Strikingly, both the gated and un-gated SLAM methods perform similarly to the gated CSI, despite both having significantly lower CRLBs than the un-gated CSI. This suggests that in both cases the effect of cardiac motion is reduced, either by the larger VOI encompassing the whole heart across the full cardiac cycle, or by this, in combination with cardiac gating. Reassuringly the correlation plots in Figure 5, which compare each of the methods to un-gated CSI show that the reduction in variation (represented by the negative gradient of the line) is equivalent for gated SLAM and gated CSI, and only slightly lower for un-gated SLAM. Hence, the reduction in variation for SLAM is genuine, and not just a result of a reduction in sensitivity caused by averaging over the expanded VOI.
Comparing the SLAM and CSI reconstructions of the main study to the HFpEF data-set (where a lower PCr/ATP ratio can be expected) highlights the experimental utility of the technique, using a two-tailed Welch’s t-test the SLAM reconstructions showed a significant difference in PCr/ATP ratio (p=0.0233) even in this small sample size, whereas the CSI reconstruction did not (p=0.1606).
Conclusion
Using the SLAM algorithm to reconstruct un-gated acquisition weighted CSI data-sets reduces the coefficient of variation by 23%, and the coefficient of repeatability by 35% in healthy controls, compared to a Fourier based reconstruction, without requiring a change to acquisition protocol.Acknowledgements
This work was supported by the Engineering and Physical Sciences Research Council (EPSRC) and Medical Research Council (MRC) (EP/L016052/1). All authors acknowledge the support of the British Heart Foundation (BHF) (FS/19/18/34252), the Oxford BHF Centre for Research Excellence (RE/13/1/30181) and the UK National Institute for Health Research (NIHR). JJM acknowledges support from a Novo Nordisk Postdoctoral Fellowship scheme run in conjunction with the University of Oxford, and also by St Hugh’s and Wadham college. LV was supported by a Sir Henry Dale Fellowship from the Wellcome Trust and the Royal Society (221805/Z/20/Z). LV also acknowledges the the Slovak Grant Agencies VEGA(2/0003/20) and APVV (19-0032).
We thank Paul Bottomley for providing code used in the implementation of the SLAM algorithm.
References
[1] S. Neubauer et al. N. Engl. J. Med. 356:1140-1151, 2007
[2] C.T.R. Rodgers et al. Magn. Reson. Med. 72:304-315, 2014
[3] Y. Zhang et al. J. Magn. Reson. 218, 66-76, 2012
[4] S. Wampl et al. Nat. Sci. Rep. 11:9268, 2021
[5] M. Kienlin et al. J. Magn. Reson. 94, 268-287,1991
[6] D.J. Tyler et al. NMR in Biomed. 22:405-413, 2009
[7] M.D. Robson et al. Magn. Reson. Med. 53:267-274, 2005
Figures

Figure 1
Top: An example segmentation for one slice of the heart, used in the SLAM reconstruction algorithm, showing (blue) chest wall, (yellow) heart, and (greyscale) other. Bottom: An example spectra, reconstructed with the SLAM algorithm, and its AMARES fit.
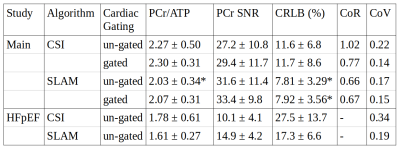
Figure 2
Mean values of the experimental results derived for each method, errors are given as ± 1 SD. * indicates significant difference to the un-gated AW CSI method (Wilcoxon signed-rank paired, α=0.05).
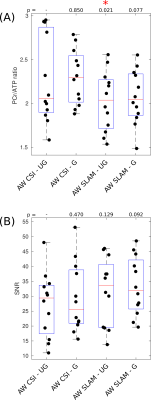
Figure 3
Box-plots showing PCr/ATP values (A) and PCR SNR (B) for each acquisition, median and IQR are indicated by box. * indicates significant difference to the un-gated CSI method (Wilcoxon signed-rank paired, α=0.05). PCr/ATP values are in the expected range.
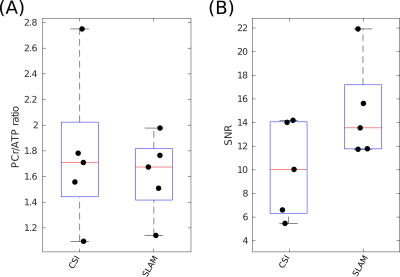
Figure 4
Box-plot showing PCr/ATP values (A) and PCR SNR (B) for each reconstruction method applied to the clinical research HFpEF data-set, median and IQR are indicated by box. PCr/ATP values are lower than the healthy control volunteers, as is expected.
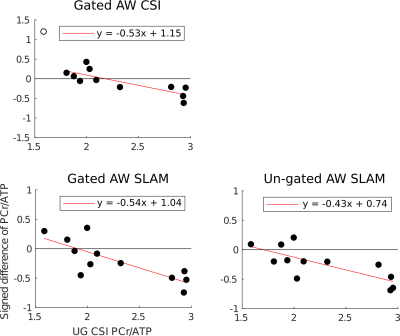
Figure 5
Correlation plots comparing each method to the un-gated CSI method. y-axis: signed difference in the PCr/ATP ratio of each method and the un-gated CSI, x-axis: PCr/ATP ratio of the un-gated CSI. An empty circle indicates the data-point was excluded from calculation of line of best fit. A negative gradient corresponds to a reduction in variation compared to the un-gated CSI. The gradient of the line is approximately equal for the gated/un-gated methods respectively.