1639
Semi-supervised learning for fast multi-compartment relaxometry myelin water imaging (MCR-MWI)1Donders Institute for Brain, Cognition and Behaviour, Nijmegen, Netherlands, 2Athinoula A. Martinos Center for Biomedical Imaging, Charlestown, MA, United States, 3Department of Radiology, Harvard Medical School, Boston, MA, United States
Synopsis
Myelin water imaging using multi-compartment relaxometry (MCR-MWI) improves the GRE-MWI robustness and accuracy but suffered from slow processing speed. In this study, we incorporate both supervised and self-supervised machine learning for fast MCR-MWI that is generalisable to a wide range of acquisition parameters without the need to re-train the network. We demonstrate its application on single compartment fitting and MCR-MWI. Results show that the proposed method can produce comparable high SNR results with a 62-fold shorter processing time.
Introduction
Gradient echo myelin water imaging (GRE-MWI) is a promising myelination measurement method, yet highly ill-conditioned1-3. A multi-compartment relaxometry method for MWI (MCR-MWI)4 incorporating variable flip angle (VFA), multi-echo GRE acquisition was recently introduced to overcome GRE-MWI limitations: accounting for distinct free water (IEW) and myelin water (MW) signal saturation, and ensuring fitting convergence. In MCR-MWI, the steady-state signal is modelled by the extended phase graph with exchange (EPG-X) framework5 to account for inter-compartmental magnetisation exchange. However, using EPG-X with non-linear least squares (NLS) fitting is computationally expensive. Without parallelisation, one 1.5-mm isotropic resolution brain volume requires ~350 computation hours.Using fully-connected artificial neural networks (FC-ANN) to speed-up parameter mapping was previously implemented for MWI6-7. However, these trained networks are protocol-specific and are not generalisable to new acquisition parameters (e.g., echo times). This would be even more critical with MCR-MWI where repetition time (TR), and (number of different) flip angles (α) have to be considered.
Self-supervised learning has been successful on rapid parameter mapping without the necessity of training data8,9. We propose a semi-supervised learning method for MCR-MWI. An FC-ANN is trained as a fast EPG-X simulator to generate the IEW and MW steady-state signal. The trained FC-ANN is then embedded in the MCR-MWI model in a self-supervised network for parameter mapping10, utilising its computational efficiency to perform optimisation for a large number of voxels without extra training data, making it applicable across a variety of acquisition settings.
Methods
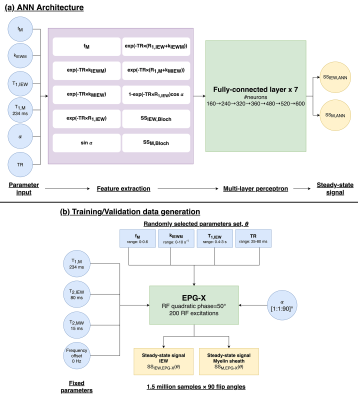
FC-ANN for EPG-XThe FC-ANN6 architecture for EPG-X is described in Fig.1. The network takes six inputs: myelin sheath volume fraction (fM), T1,IEW, T1,M, exchange rate (kIEWM), α, and TR, and returns the magnitude steady-state IEW and MW signals that match the EPG-X simulations.
Training data was generated from 1.5x106 random parameter sets (θ) with the 4 parameters described in Fig.1b. Training was performed using Adam optimiser with 100 epochs. The training loss was defined as a sum of three mean squared errors (MSE):
$$loss=MSE_{\theta,\alpha}+MSE_{\theta,1-90}+{\lambda}MSE_{ds_\theta/d\alpha}$$
corresponding to the MSE between the FC-ANN predictions and the EPG-X signal (1) given θ at a single α ($$$MSE_{\theta,\alpha}$$$) and (2) for all 90 flip angles ($$$MSE_{\theta,1-90}$$$), and (3) the first derivative of the signal to flip angle ($$$MSE_{ds_\theta/d\alpha}$$$).
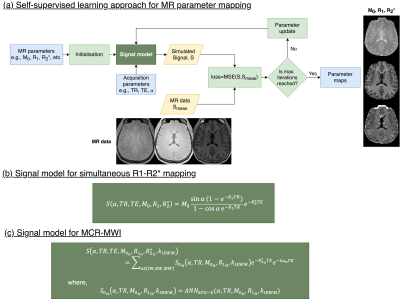
Self-supervised learning for parameter mapping
To perform parameter mapping, we deployed a simplistic physics-informed self-learning approach10 (Fig.2). The network is initialised with random (and/or constant) values for the MR parameters (which are the network parameters to be optimised) and the acquired MR data is given as input. During the learning process, signal is simulated given the parameters and the signal model (Fig.2b,c), and the MR parameters are updated using an Adam optimiser based on a loss function of the MSE between simulated and actual signal.
Both networks were created and trained using the Deep Learning Toolbox in Matlab R2021b (Natwick, US) with an NVIDIA Tesla P100 GPU (Santa Clara, CA).
In vivo imaging
Data acquisition was performed at 3T (Siemens, Erlangen) on 2 healthy volunteers. A monopolar 3D ME GRE sequence was used to acquire VFA data using two distinct sets of protocols
1) TR/TE1/ΔTE/nTE=38/2.2/3.07ms/12, TA=2.8min/α;
2) TR/TE1/ΔTE/nTE =55/2.68/3.95ms/13, TA=4min/α,
res=1.5mm iso., α=[5,10,20,50,70]°, RCAIPI=5. B1 map was acquired to correct the B1 field inhomogeneity. The complex data of all different α datasets and the B1 map were co-registered before further processing.
Single Compartment Relaxometry
As a reference standard fast processing pipeline, R2* was estimated on each dataset using trapezoidal integration11, followed by DESPOT1 R1 mapping on the extrapolated S0 images12. Mean R2* map was computed across flip angles. The self-supervised learning method shown in (Fig.2a,b) was used to demonstrate the network ability to perform simple parameter mapping.
Multi-Compartment Relaxometry
MWF maps were obtained as in 4 using an NLS fitting on a voxel-wise basis and compared to the self-supervised learning method described in (Fig.2a,c), processing ~12000 voxels simultaneously per batch.
The resulting maps between standard and self-supervised data were compared.
Results and Discussion
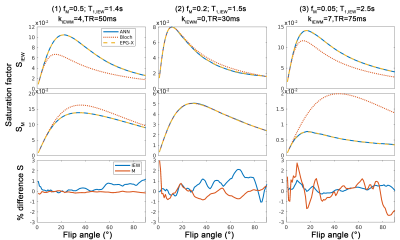
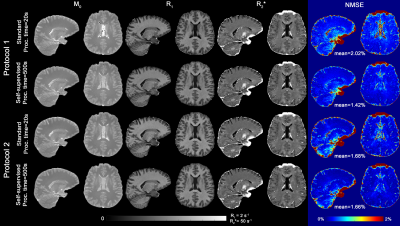
Fig.3 shows that the FC-ANN can generate the EPG-X signal for a variety of protocols and tissue parameters with the maximum percentage difference being below 3%.Fig. 4 shows that the M0, R1* and R2* maps derived from self-learning and standard relaxometry deliver comparable results, but the normalised MSE between the simulated and measured data is lower with the self-supervised method benefiting from its explicit MSE cost function.
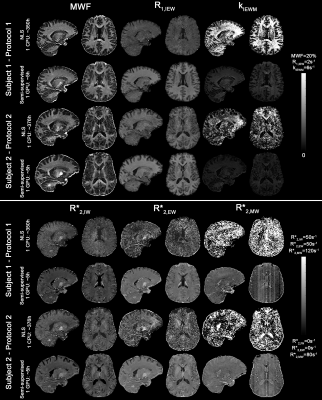
Fig. 5 shows that the semi-supervised approach results in comparable MWF maps to voxel-wise fitting but 62-fold faster. Banding artefacts are observed in the most SNR sensitive measurements (R2,MW* and kIEWM) in regions corresponding to different processing batches. Although no explicit spatial regularization was used, the maps obtained are less prone to noise enhancement which is attributed to the learning gradients being computed over a large number of pixels.
Conclusions
We present a semi-supervised framework for MCR-MWI using an FC-ANN that is flexible to protocol settings (TR, TE and α) without re-training. Future work will explore the impact of learning rates associated with the various maps and introduce a 3DTV loss function to allow higher resolution MCR-MWI protocols. This framework can be adapted to complex non-linear fitting approaches as quantitative CEST or MT.Acknowledgements
This work is part of the research programme with project number FOM-N-31/16PR1056/RadboudUniversity, which is financed by the Netherlands Organisation for Scientific Research (NWO). BB and THK are supported by research grants NIH R01 EB028797, R03 EB031175, U01 EB025162, P41 EB030006, U01 EB026996, and the NVidia Corporation for computing support.
References
1. Nam Y, Lee J, Hwang D, Kim D-H. Improved estimation of myelin water fraction using complex model fitting. Neuroimage 2015;116:214–221.2. Alonso-Ortiz E, Levesque IR, Pike GB. Impact of magnetic susceptibility anisotropy at 3 T and 7 T on T2*-based myelin water fraction imaging. Neuroimage 2018;182:370–378.
3. Lee J, Nam Y, Choi JY, Kim EY, Oh S-H, Kim D-H. Mechanisms of T2 * anisotropy and gradient echo myelin water imaging. NMR in biomedicine 2016.
4. Chan K-S, Marques JP. Multi-compartment relaxometry and diffusion informed myelin water imaging – Promises and challenges of new gradient echo myelin water imaging methods. Neuroimage 2020;221:117159.
5. Malik SJ, Teixeira RPAG, Hajnal JV. Extended phase graph formalism for systems with magnetization transfer and exchange. Magnetic resonance in medicine 2017;3:125 doi: 10.1002/mrm.27040.
6. Lee J, Lee D, Choi JY, Shin D, Shin H-G, Lee J. Artificial neural network for myelin water imaging. Magnetic resonance in medicine 2019;31:673.
7. Jung S, Lee H, Ryu K, et al. Artificial neural network for multi‐echo gradient echo–based myelin water fraction estimation. Magnet Reson Med 2021;85:380–389.
8. Kang B, Kim B, Schär M, Park H, Heo H. Unsupervised learning for magnetization transfer contrast MR fingerprinting: Application to CEST and nuclear Overhauser enhancement imaging. Magnet Reson Med 2021;85:2040–2054.
9. So S, Kim B, Park H, Bilgic B. BUDA-STEAM: A rapid parameter estimation method for T1, T2, M0, B0 and B1 using three-90° pulse sequence. In: Processing
30, Annual Meeting International Society for Magnetic Resonance in Medicine, Montreal, Canada, 0327 (2021).
10. Kim T, Cho J, Zhao B, Bilgic B. MR parameter mapping with unsupervised scan-specific neural networks. Workshop on MRI Acquistion & Reconstruction, Virtual meeting (2021)
11. Gil R, Khabipova D, Zwiers M, Hilbert T, Kober T, Marques JP. An in vivo study of the orientation-dependent and independent components of transverse relaxation rates in white matter. NMR in biomedicine 2016;29:1780–1790.
12. Deoni SCL, Rutt BK, Peters TM. Rapid combinedT1 andT2 mapping using gradient recalled acquisition in the steady state. Magnet Reson Med 2003;49:515–526.
Figures