1589
Towards Automated Scan Volume Placement for Breast MRI Using a Deep Neural Network1Medical Physics, University of Wisconsin-Madison, Madison, WI, United States, 2Radiology, University of Wisconsin-Madison, Madison, WI, United States, 3GE Healthcare, Houston, TX, United States, 4GE Healthcare, Waukesha, WI, United States, 5GE Healthcare, Menlo Park, CA, United States, 6GE Healthcare, Madison, WI, United States, 7Radiology, University of Iowa, Iowa City, IA, United States
Synopsis
We investigate the use of a deep neural network to automatically position imaging volumes for breast MR exams. The axial localizer images and scan volume information from a variety of MR scanners were used to train a deep neural network to replicate the clinical technologists’ placement. The average intersection over union between clinical placement and neural network predicted placement was 0.46 ± 0.21. The distance between volume centers was 7.4 cm on average and as low as 1.1 cm. These results show promise for improving consistency of imaging volume placement in breast MRI.
Introduction
Abbreviated breast protocols have shown great promise and efforts to date have primarily focused on optimizing the imaging protocols1. However, efficiency gains in the exam setup have seen little investigation. In recent years, a limited number of deep learning solutions for automated placement of MR scan volumes have been introduced with applications including neuro, cardiac, and liver volumes2,3,4. The potential benefits of robust scan volume placement include greater reproducibility, reduced workload for technologists, a reduction in exams with inadequate coverage, and potential time saving based on the speed of the placement algorithm. The timing benefits would be particularly pronounced if used in conjunction with automated local shim volume placement as previously investigated by Kang et al5. Breast exam setup includes 3D visualization of the spatial overlap of anatomy, scan volume, and local shim volumes with typical workflows rely on confirming positioning in all 3 planes of localizer images. The complicated workflow is time consuming and mentally taxing for MR technologists contributing several minutes to the total table time in our experience.Automated FOV and shim volume placement via deep learning has promise due to the complex and individually varying patient anatomy found in breast MR. Once trained, neural networks are quick to make predictions and could potentially reduce graphical prescription time to under one minute while providing consistent placement and relieving technologist burden. Eventually, this technology may see multiple neural networks used for each aspect of scan prescription (scan volume and shim volumes).
In this work, we aim to develop a deep learning-based algorithm for placement of the scan volume in breast MRI using axial localizer images as input. We hypothesis that the algorithm will be able to predict the scan volume matching those placed by trained technologists.
Methods
A total of 413 breast MR exams were collected from our institution’s PACS as part of this retrospective, IRB approved, HIPAA compliant study. Breast patients were scanned for clinical purposes on 1.5T or 3.0T scanners (GE Healthcare, Waukesha WI, see Table 1) using dedicated breast coils. Exams containing silicone or saline implants, patients who had undergone mastectomy, exams missing localizer images, or missing scan volume prescription were then excluded, leaving 328 exams. Axial localizer images and locations of the imaging volumes were extracted. The placement information was characterized by a 5 parameter vector (x,y,z position, transverse size, and longitudinal extent; all in cm). Localizer images were normalized and linearly interpolated so the through-plane resolution matched in-plane resolution (1.5-1.7 mm). To prevent overtraining, data augmentation was performed by adding Gaussian pixel noise and image shifts.A deep convolutional neural network was implemented in Keras with a TensorFlow backend6,7, consisting of a 3-D framework with four convolutional layers and two fully connected layers. It takes as input the 3-D stack of interpolated axial localizer images and predicts the 5 scan volume placement parameters. The loss function used for training was the root-mean-squared error (RMSE) between the predicted, p, and known, k, placement parameter vectors defined as
$$RMSE = \sqrt{\overline{(\textbf{p} -\textbf{k})^2}}.$$
80% of the data were used for training and 20% reserved for validation. The predicted placement was compared to the clinical technologists’ placement using three metrics: RMSE, distance between volume centers, and intersection over union (IoU) as defined in Figure 1. An IoU of 1 was considered to be perfectly aligned and 0 is considered no alignment. Time required for prediction was measured for ten of the validation exams.
Results
Examples of the predicted scan volumes from the deep learning model can be found in Figure 2 alongside their respective RMSE and IoU values. The average RMSE obtained over the validation dataset was 5.2 ± 4.2 cm (max: 27.4, min: 0.6) while the average IoU was 0.46 ± 0.21 (max: 0.83, min: 0.0). The distance between volume centers was 7.4 cm on average and as low as 1.1 cm (max: 32.6). Figure 3 shows distributions of both RMSE and IoU for the entire validation dataset. Figure 4 shows variation of the estimation error for each of the individual placement parameters. The average time for volume prediction was 14.7 seconds.Discussion and Conclusions
Although this feasibility study was performed in a relatively limited dataset, the neural network was capable of IoUs up to 0.83 indicating good overlap between the model predictions and the clinical technologists’ pre-scan volumes. In terms of RMSE, 2 outlier cases were observed. Visual inspection of these subjects reveals large breast size with one of the subjects having the breast tissue deformed by the scanning table. Additional training data with larger breast sizes is expected improve performance. The current study uses a relatively limited dataset and does not yet fully encompass the wider range of body geometries including more complex cases such as mastectomy and implants.We demonstrated a deep neural network for automatic prediction of scan volume placement in breast MR. With further training and in conjunction with a similar algorithm for shim volume placement, full automation of the graphical scan prescription for breast MR is possible with benefits of consistent scan/shim placement, simplified technologist workflow, and reduced scan setup time.
Acknowledgements
University of Wisconsin receives funding from GE Healthcare. This project was supported by NIH 1R01CA248192 and the Departments of Radiology and Medical Physics, University of Wisconsin.References
1. Blansit, K., Retson, T., Masutani, E., Bahrami, N. & Hsiao, A. Deep Learning–based Prescription of Cardiac MRI Planes. Radiology: Artificial Intelligence 1, e180069 (2019).
2. Shanbhag, D et al. A Generalized Deep Learning Framework for Multi-landmark Intelligent Slice Placement Using Standard Tri-planar 2D Localizers. ISMRM Annual Meeting 2019 (2019).
3. Geng, R et al. Automated Image Prescription for Liver MRI using Deep Learning. ISMRM Annual Meeting 2021 (2021).
4. Wang K et al. Toward Fully Automated Breast MR Exams using Deep Learning. ISMRM Annual Meeting 2018 (2018)
5. Kuhl CK, Schrading S, Strobel K, Schild HH, Hilgers RD, Bieling HB. Abbreviated breast magnetic resonance imaging (MRI): first postcontrast subtracted images and maximum-intensity projection: a novel approach to breast cancer screening with MRI. J Clin Oncol 2014;32(22):2304–2310.
6. https://keras.io/
7. https://www.tensorflow.org/
Figures
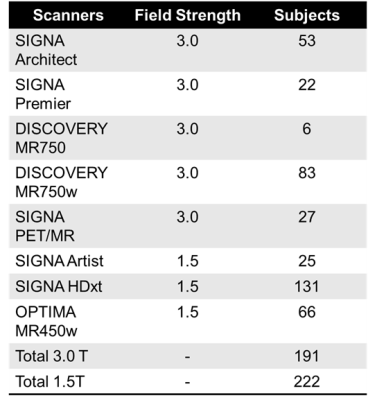
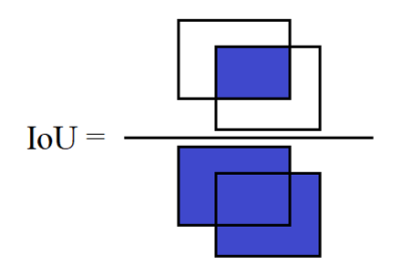
Figure 1: Definition of the intersection over union (IoU) measuring the overlap between two scan volumes. The IoU is calculated as the intersection of the two volumes divided by the union of the two volumes. Note that three-dimensional volumes were used in this abstract.
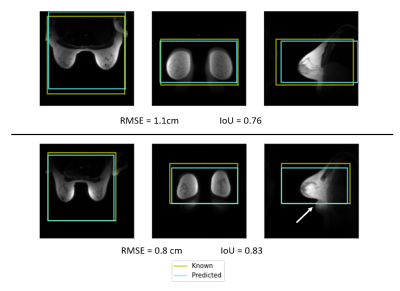
Figure 2: Example scan volume placements for two subjects showing both the scan volume placed by the clinical technologist (yellow) and the deep-learning prediction (blue) in the axial, coronal, and sagittal planes. These two examples show excellent overlap between the two scan volumes. The lower example shows tissue bunched under the breast (arrow) and the DL model was able to correctly predict placement despite the bunched tissue.
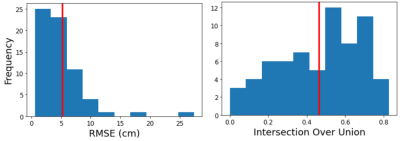
Figure 3: Distributions of the RMSE and IoU metrics over the validation dataset showing the variation in scan volume placement accuracy. Vertical red lines indication mean values.
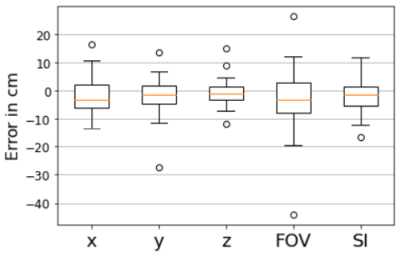
Figure 4: Boxplots showing the variation of the error in the individual imaging volume placement parameters. Positional information (x,y,z) generally varied between -15 to 15 cm with a few outliers (circles). FOV represents the size in the axial plane and showed the most variation and the outliers with the greatest error. SI is the extent of the imaging volume in the SI direction.