1579
Deep learning voxelwise classification of primary central nervous system lymphoma using DSC-PWI normalized time-intensity curves1Vall d'Hebron Institute of Oncology, Barcelona, Spain, 2Bellvitge University Hospital, L'Hospitalet de Llobregat, Spain, 3Hospital Clínic de Barcelona, Barcelona, Spain
Synopsis
The robustness and accuracy of current MR methods to differentiate brain tumours is limited. In this study we investigate the potential of dynamic susceptibility perfusion-weighted imaging (DSC-PWI) normalized time-intensity-curves (nTIC) to support lymphoma diagnosis by harnessing voxelwise and temporal information to train a convolutional neural network (CNN). This novel approach discriminated patients with lymphoma from glioblastoma and metastasis with an average accuracy of 0.94, using only a limited number of patients for training, outperforming standard DSC-PWI measurements. Furthermore, it provides voxel-by-voxel lymphoma probability maps to further help visual diagnosis of neuroradiologists.
Introduction
Presurgical non-invasive differential diagnosis of enhancing brain tumours in morphological MR imaging is challenging1,2 and significantly impacts patient management, i.e. suspected glioblastomas may undergo maximal safe resection, while not being the treatment of choice for a lymphoma suspicion. Dynamic susceptibility contrast- perfusion weighted imaging (DSC-PWI) consists of a temporal T2* acquisition during the vascular pass of a contrast bolus which provides insight into tissue vascularization. DSC-PWI parameters have been reported useful for brain tumour differentiation. However, heterogeneity in the applied parameters and thresholds prevents robust generalized methods of tumour classification3-7. This issue is approached by a recent method to normalize time-intensity-curves (nTICs) that allows whole nTIC unsupervised analysis of heterogeneous samples8. We present a new deep learning (DL) approach based on voxelwise nTIC convolutional neural networks (CNN) for a more accurate diagnosis of lymphoma versus the two most common brain tumours: glioblastoma and metastases.Methods
DataData from patients with histologically-confirmed brain glioblastoma, primary central nervous system lymphoma or solitary metastasis were retrospectively analysed. DSC-PWI and contrast-enhanced T1-weighted (CE-T1W) images were acquired before any treatment.
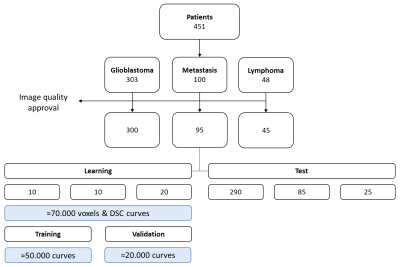
DSC-PWI were acquired with 1.5T Philips Ingenia and Intera scanners. Salient sequence (gradient-echo EPI and PRESTO9) parameters were: temporal sampling of 1.26 to 1.93 seconds, 40 to 60 timepoints, resolution of 1.5 and 1.7mm, TR/TR2 of 1640 and 16ms. A total of 451 patients from a single institution were collected: glioblastoma (n=303), metastasis (n=100) and lymphoma (n=48) (Figure 1).
Post-processing
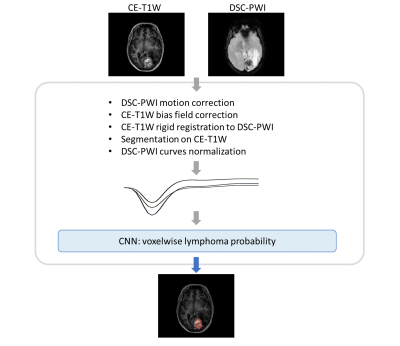
Scans were visually approved by an experienced neuroradiologist and processed as shown in Figure 2. DSC-PWI curves were normalized8 to obtain nTICs. On the CE-T1W, brain masking was performed with a hierarchical approach10, enhancing tumour and white matter were segmented by thresholding and revised by a neuroradiologist.
DL training and classification
Patients were randomly split into equally-sized groups for training (20 patients with lymphoma, 10 with glioblastoma and 10 with metastasis), while 25 patients with lymphoma, 85 with metastasis and 290 with glioblastoma were used for testing (balanced for acquisition sequences).
Using every voxel of the enhancing tumour, 69354 nTICs were used to train a 1D CNN for identifying lymphoma curves (Figure 1). The CNN provides voxelwise spatial maps within the enhancing tumour, indicating each voxel’s probability of exhibiting a lymphoma-like perfusion (Figure 3), while low probabilities correspond to other tumours (glioblastoma/metastasis). The ratio of voxels with a probability higher than 0.9 (lymphoma) over voxels with probability higher than 0.9 and lower than 0.1 (other) was used to classify a patient.
The new DL classification was benchmarked against logistic regression with mean relative cerebral blood volume (rCBV) and mean percentage of signal recovery (PSR). Classification thresholds were obtained by Youden’s index from the training set.
The area under the receiver-operating characteristic curve (ROC AUC) was obtained and 100 groups of 25 random test patients were used to obtain the classifying performance average and 95% confidence intervals (CI).
Results
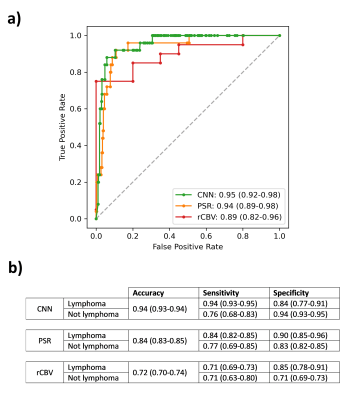
The CNN of voxel-by-voxel nTICs identified patients with lymphoma in the test set with a ROC AUC of 0.95 (CI: 0.92-0.98) and an accuracy of 0.94 (CI: 0.93-0.94) when classifying 100 random test groups. Mean rCBV obtained an AUC of 0.89 (CI: 0.82-0.96) and an accuracy of 0.72 (CI: 0.70-0.74) for the 100 groups. Mean PSR classified patients with a ROC AUC of 0.94 (CI: 0.89-0.98) and an accuracy of 0.84 (CI: 0.83-0.85) in the random test groups. The logistic regression combining mean rCBV and PSR obtained an AUC of 0.91 (CI: 0.84-0.98) and an accuracy of 0.80 (CI: 0.78-0.81) for the 100 groups (Figure 4).Discussion
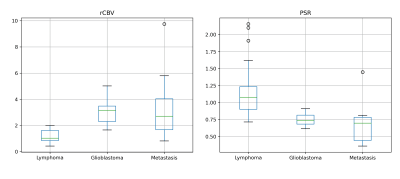
A CNN was successfully trained to identify patients with lymphoma, outperforming standard perfusion markers. Our approach had higher accuracy and narrower confidence intervals than rCBV and PSR. With Youden’s index, the CNN showed high sensitivity for lymphoma and high specificity for other tumours. This may be clinically relevant, e.g.: skipping biopsies currently needed to rule out lymphoma.The heterogeneity of rCBV and PSR across publications5,6 make it difficult to find absolute references (Figure 5). Combining rCBV and PSR did not improve the classification, supporting the use of all curve timepoints to capture the DSC-PWI potential, as proposed by our group8.
A prior publication using CNNs11 uses only five parameters of the dynamic curves and represents both lymphoma and glioblastoma with the same DSC-PWI curve patterns from tumour subregions. We hypothesized that tumour differentiation can be approached considering all timepoints of the curves and the whole enhancing tumour. Additionally, the probability colourmaps may be of clinical interest (e.g. guiding biopsies) and provide user-friendly visual outcomes.
Potential limitations to address include testing the method on an external cohort and studying segmentation variability effects on performance. As future work, we shall also extend the differentiation to other tumour types.
Conclusion
We present a new method of analysing DSC-PWI nTICs taking advantage of all voxels and all curve timepoints to identify patients with lymphoma from glioblastoma and metastasis. Our results outperform conventional perfusion parameters and provide spatial probability maps, which could inform visual inspections of neuroradiologists. Moreover, our framework based on DSC-PWI nTIC voxelwise CNN overcomes the lack of homogeneity in perfusion protocols and enables robust DL classification with just few patients, which has therefore strong clinical appeal.Acknowledgements
This project has received support from “la Caixa” Foundation (RTI2018-095209-B-C21), Spanish Ministry of Science and Innovation (FIS-G64384969). FG is supported by the investigator-initiated PREdICT study at the Vall d'Hebron Institute of Oncology (Barcelona), funded by AstraZeneca. RPL is supported by a CRIS Foundation Talent Award (TALENT19-05), the Instituto de Salud Carlos III-Investigación en Salud (PI18/01395) and the Prostate Cancer Foundation Young Investigator Award. None of these influenced data acquisition, analysis and result interpretation.References
1. Leung D, Han X, Mikkelsen T, Nabors LB. Role of MRI in primary brain tumor evaluation. J Natl Compr Canc Netw 2014;12(11):1561-1568.
2. Arita K, Miwa M, Bohara M, Moinuddin FM, Kamimura K, Yoshimoto K. Precision of preoperative diagnosis in patients with brain tumor - A prospective study based on "top three list" of differential diagnosis for 1061 patients. Surg Neurol Int 2020;11:55.
3. Cha S, Lupo JM, Chen MH, et al. Differentiation of glioblastoma multiforme and single brain metastasis by peak height and percentage of signal intensity recovery derived from dynamic susceptibility-weighted contrast-enhanced perfusion MR imaging. AJNR Am J Neuroradiol 2007;28(6):1078-1084.
4. Mangla R, Ginat DT, Kamalian S, et al. Correlation between progression free survival and dynamic susceptibility contrast MRI perfusion in WHO grade III glioma subtypes. J Neurooncol 2014;116(2):325-331.
5. Mangla R, Kolar B, Zhu T, Zhong J, Almast J, Ekholm S. Percentage signal recovery derived from MR dynamic susceptibility contrast imaging is useful to differentiate common enhancing malignant lesions of the brain. AJNR Am J Neuroradiol 2011;32(6):1004-1010.
6. Lee MD, Baird GL, Bell LC, Quarles CC, Boxerman JL. Utility of Percentage Signal Recovery and Baseline Signal in DSC-MRI Optimized for Relative CBV Measurement for Differentiating Glioblastoma, Lymphoma, Metastasis, and Meningioma. AJNR Am J Neuroradiol 2019;40(9):1445-1450.
7. Surendra KL, Patwari S, Agrawal S, Chadaga H, Nagadi A. Percentage signal intensity recovery: A step ahead of rCBV in DSC MR perfusion imaging for the differentiation of common neoplasms of brain. Indian J Cancer 2020;57(1):36-43.
8. Pons-Escoda A, Garcia-Ruiz A, Naval-Baudin P, et al. Presurgical Identification of Primary Central Nervous System Lymphoma with Normalized Time-Intensity Curve: A Pilot Study of a New Method to Analyze DSC-PWI. AJNR Am J Neuroradiol 2020;41(10):1816-1824.
9. van Gelderen P, Duyn JH, Ramsey NF, Liu G, Moonen CT. The PRESTO technique for fMRI. Neuroimage 2012;62(2):676-681.
10. Pohl KM, Bouix S, Nakamura M, et al. A hierarchical algorithm for MR brain image parcellation. IEEE Trans Med Imaging 2007;26(9):1201-1212.
11. Park JE, Kim HS, Lee J, et al. Deep-learned time-signal intensity pattern analysis using an autoencoder captures magnetic resonance perfusion heterogeneity for brain tumor differentiation. Sci Rep 2020;10(1):21485.
Figures