1436
Exploring Hybrid CNN-Transformer for Schizophrenia Classification using Structural MRI1Department of Biomedical Engineering, Columbia University, New York, NY, United States, 2Department of Psychiatry, Columbia University, New York, NY, United States
Synopsis
Schizophrenia diagnosis is clinically difficult due to the lack of biomarkers associated with the disease. While machine learning algorithms and convolutional neural networks (CNNs) have found success using neuroimaging inputs to diagnose the disease, they have historically not performed or generalized well enough for clinical use. We propose 3D-MIC-Transformer, the first transformer-based deep learning architecture applied to neurological disease classification that demonstrates state-of-the-art schizophrenia classification performance and generalization using structural MRI inputs. 3D-MIC-Transformer outperforms prior CNN implementations (AUROC: 0.985, accuracy: 0.933), and we believe 3D-MIC-Transformer can serve as a backbone for other disease classification tasks in the future.
Introduction
Diagnosis of schizophrenia is difficult because there does not currently exist a biomarker indicating the onset of the disease1. While trained clinicians can identify schizophrenia post disease progression, symptoms often overlap with other mental disorders necessitating careful analysis to differentiate between them. Magnetic Resonance Imaging (MRI) presents the ability to observe anatomical differences indicative of schizophrenia development2,3, making them ideal inputs for schizophrenia screening tools.Machine learning algorithms such as support vector machines have been historically successful using MRI, but their performance largely depends on the quality of manually extracted features4,5. Alternatively, while convolutional neural networks (CNNs) have recently achieved impressive performance due to their feature encoding capabilities, they often struggle to generalize well to new datasets6-7. As such, there remains a great need for an accurate and generalized schizophrenia screening tool.
Transformers have recently emerged as a new deep learning alternative that have outperformed CNNs on various tasks including disease classification8. In this study, we introduce 3D Medical Image Classification Transformer (3D-MIC-Transformer), a CNN-transformer hybrid deep learning (DL) architecture that achieves state-of-the-art schizophrenia classification performance and generality.
Methods
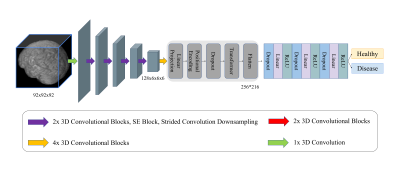
The architecture for the proposed model is shown in Figure 1. 3D-MIC-Transformer consists of a 5-layered CNN encoder with squeeze-excitation (SE) blocks9 before each down-sampling operation. The encoder portion is followed by a transformer module, which then feeds into a three-layer fully-connected classifier module. We compared 3D-MIC-Transformer to the prior state-of-the-art schizophrenia classification implementation described in Oh et al6. Moreover, we also compared 3D-MIC-Transformer to a VGG-11BN 3D-CNN classifier that has been successfully applied to Alzheimer’s classification10,11, which we further improved by introducing SE blocks. Both 3D-MIC-Transformer and SE-VGG-11BN were trained using 3D downsampled 92x92x92 MRI inputs for 300 epochs and optimized using Adam for the first 100 epochs and with SGD afterwards. The benchmark model was trained following the protocol outlined in Oh et al6.We collected data from BrainGluSchi12, COBRE13, and NMorphCH14 from the SchizConnect database. The acquisition parameters and characteristics for each dataset are outlined in Figure 2a while the pre-processing pipeline is described in Figure 2b. We first trained and tested 3D-MIC-Transformer, SE-VGG-11-BN, and the benchmark model on the same train/validation/test (8:1:1) split using the combined BrainGluSchi, COBRE, and NMorphCH datasets. We trained using early stopping based on the best validation performance. We then compared the generality between the models by training/validating them on COBRE/NMorphCH and testing them on the unseen BrainGluSchi dataset. We quantified model performance using the area under the ROC curve (AUROC) metric along with accuracy, sensitivity, and specificity. We tested for significant AUROC differences between our models and the baseline model using DeLong’s test.
Github Repository for 3D-MIC-Transformer can be found at: https://github.com/raovish6/3D-MIC-Transformer
Results
Both 3D-MIC-Transformer and SE-VGG-11BN outperformed the prior state-of-the-art implementation on the first train/validation/test experiment, with significantly higher AUROC scores (p≤0.05). Additionally, 3D-MIC-Transformer achieved higher AUROC, accuracy, and specificity compared to the benchmark model and SE-VGG-11BN, with a max AUROC score of 0.985 and accuracy of 0.933. These results are shown in Figure 3.We also found that 3D-MIC-Transformer and SE-VGG-11BN generalized better than the implementation from Oh et al6, with significantly AUROC performance (p≤0.05). Moreover, 3D-MIC-Transformer exhibited higher AUROC, accuracy, and sensitivity compared to the benchmark model and SE-VGG-11BN. These results are shown in Figure 4.
Discussion
This study investigated the performance of a new CNN-transformer hybrid schizophrenia classification model. 3D-MIC-Transformer showcased superior performance and generalization compared to the prior state-of-the-art implementation. Moreover, 3D-MIC-Transformer also outperformed a separate improved 3D-CNN implementation designed for disease classification.Our model represents the first successful application of transformers to neurological disease classification using MRI data. Transformers present the ability to capture global contextual information and long-distance interdependencies in the input data that are often missed by the local receptive fields of CNNs. By incorporating both CNN and transformer encoding components into our model, we are able to take advantage of both streams of information for a more accurate and generalized classification. While SE-VGG-11BN performed only slightly worse than 3D-MIC-Transformer on our first experiment, we observed larger differences on our generalization testing which is where CNNs usually struggle. Our results indicate that the introduction of the transformer represents an efficient method of improving model performance and more importantly model generalization. Therefore, our architecture offers a universal framework for all neuroimaging based classification pipelines. The improved generalization of our model also makes it preferable for real-world applications.
While our model reaches state-of-the-art performance, we believe it can be further improved by taking both structural and functional information into account as done in Zhu et al15. Additionally, we used a relatively small sample size for 3D CNN training, and our model would likely improve when trained on a larger dataset.
Conclusion
In this study, we propose 3D-MIC-Transformer, a new CNN-transformer hybrid DL model achieving state of the art schizophrenia classification performance and generality. We believe our architecture can be successfully applied to related disease classification tasks in the future.Acknowledgements
This work was supported by and performed at Zuckerman Mind Brain Behavior Institute MRI Platform, a shared resource, and Columbia MR Research Center site.References
Kraguljac NV, McDonald WM, Widge AS, et al. Neuroimaging biomarkers in schizophrenia. American Journal of Psychiatry. 2021; 178(6):509–521.
Koutsouleris N, Riecher-Rössler A, Meisenzahl EM, et al. Detecting the psychosis prodrome across high-risk populations using neuroanatomical biomarkers. Schizophrenia Bulletin. 2015; 41(2):471–482.
Patel KR, Cherian J, Gohil K, et al. Schizophrenia: overview and treatment options. P&T. 2014; 39(9):638–645
Arbabshirani MR, Plis S, Sui J, et al. Single subject prediction of brain disorders in neuroimaging: Promises and pitfalls. Neuroimage. 2017; 145:137–165
Davatzikos C. Machine learning in neuroimaging: Progress and challenges. Neuroimage. 2019; 197:652.
Oh J, Oh Baek-Lok, Lee K, et al. Identifying schizophrenia using structural mri with a deep learning algorithm. Frontiers in Psychiatry. 2020; 11:16.
Hu M, Qian X, Liu S, et al. Structural and diffusion MRI based schizophrenia classification using 2D pretrained and 3D naive Convolutional Neural Networks. Schizophrenia Research. 2021.
Dai Y, Gao Y, Liu F. TransMed: Transformers Advance Multi-Modal Medical Image Classification. Diagnostics. 2021; 11(8):1384.
Hu J, Shen L, and Sun G. Squeeze-and-Excitation Networks. CVPR. 2018; 7132-7141.
Zhu N, Liu C, Feng X, et al. Deep Learning Identifies Neuroimaging Signatures Of Alzheimer’s Disease Using Structural And Synthesized Functional Mri Data. ISBI. 2021; 216-220.
Feng X, Yang J, Lipton Z, et al. Deep Learning on MRI Affirms the Prominence of the Hippocampal Formation in Alzheimer's Disease Classification. bioRxiv. 2018; 10.1101/456277.
Chyzhyk D, Savio A, and Graña M. Computer aided diagnosis of schizophrenia on resting state fMRI data by ensembles of ELM. Neural Networks. 2015; 68:23-33.
Bustillo JR, Jones T, Chen H, et al. Glutamatergic and neuronal dysfunction in gray and white matter: a spectroscopic imaging study in a large schizophrenia sample. Schizophr Bull. 2017; 43(3):611-619.
Alpert K, Kogan A, Parrish T, et al. The northwestern university neuroimaging data archive (NUNDA). NeuroImage. 2016; 124(B):1131-1136.
Zhu N, Liu C, Feng X, et al. Deep learning identifies neuroimaging signatures of Alzheimer’s disease using structural and synthesized functional mri data. ISBI. 2021; 216-220
Vaswani A, Shazeer N, Parmar N, et al.. Attention Is All You Need. NeurIPS. 2017; 30
Figures
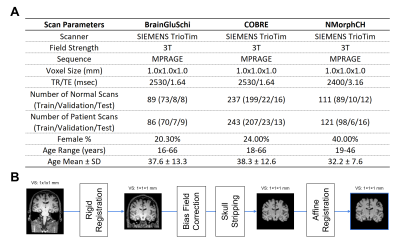
A. Acquisition and scanner parameters for each of the three primary datasets evaluated. All images were obtained from 3T MPRAGE scanners with the same voxel size and similar TR/TE, allowing us to merge them in our experiments. B. Pre-processing pipeline for MRI scans used to train and test the 3D-MIC-Transformer and SE-VGG-11BN models. This process involved rigid registration, bias field correction, skull stripping, and affine registration as visualized in the figure.
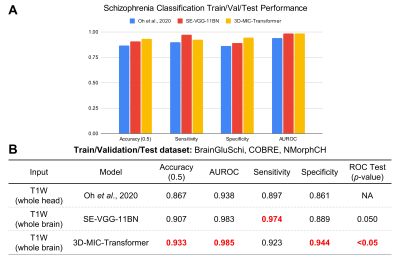
A. Bar graph visualizing the train/validation/test results compared across the three models. B. Table summarizing the schizophrenia train/validation/test classification performance metrics across the three models tested, with red boldings indicating superior performance. Both 3D-MIC-Transformer and SE-VGG-11BN had significantly higher AUROC scores compared to the benchmark model. 3D-MIC-Transformer achieved better accuracy, AUROC, and specificity compared to the other models.
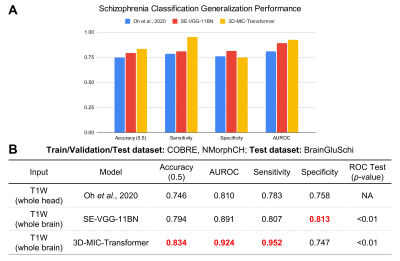
A. Bar graph visualizing the generalization performance compared across the three models. B. Table summarizing the schizophrenia classification generalization performance metrics across the three models tested, with red boldings indicating superior performance. Both 3D-MIC-Transformer and SE-VGG-11BN had significantly higher AUROC scores compared to the benchmark model. 3D-MIC-Transformer achieved better accuracy, AUROC, and sensitivity compared to the other models.