1430
Multi-Channel Deep Learning for IDH Mutation Status Prediction in Gliomas:A Multimodal Approach1Department of Magnetic Resonance, LanZhou University Second Hospital, Lanzhou, China, 2Vipshop (China) Co. LTD, Guangzhou, China
Synopsis
Isocitrate dehydrogenase (IDH) mutation status has emerged as an important prognostic marker in gliomas.Currently, reliable IDH mutation determination requires invasive surgical procedures. Various studies have demonstrated the efficacy of deep learning in classify IDH status using Magnetic resonance images(MRI)data.In this study we propose a multi-channel architecture of 3D convolutional neural networks (CNNs) based on deep learning to predict the IDH status.We utilize traditional structures imaging and various diffusivity metric maps derived from diffusion tensor imaging (DTI) as input to the network.The final model achieved the AUC value of 0.93.
Introduction
One of the most important recent discoveries in glioma biology has been the identification of IDH mutation status as a marker of treatment and prognosis[1-2]. IDH mutants have a better prognosis and treatment outcome than non-mutated or wild-type gliomas[3]. Currently, the only reliable way to determine IDH mutation status is to obtain glioma tissue by invasive brain biopsy or open surgical resection[4]. The ability to develop non-invasive methods with high accuracy to determine IDH mutation status is important for determining treatment and predicting prognosis[5]. Here, we propose a highly accurate deep learning network that, by combining traditional structural imaging and diffusion tensor imaging, can better judge the IDH mutation status of glioma patients than previously published methods, helping clinical decisions on treatment and prognosis.Methods
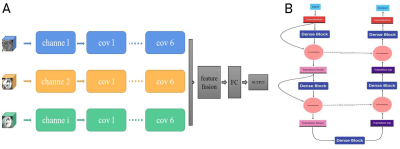
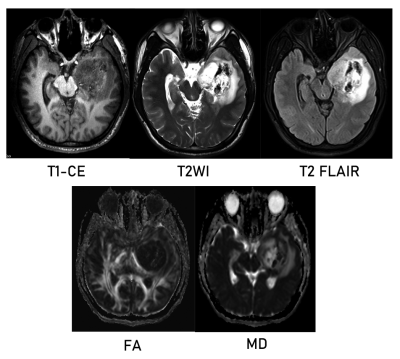
This study included 76 patients with high-grade glioma screened by preoperative imaging from the glioma image database (collected between 2016 and 2020) of Lanzhou University Second Hospital, China.This dataset called training dataset. We also included another dataset as independent data set of patients from the TCIA database to further validate our model,which with all four key sequences(T1-CE MRI /T2 MRI/ T2-FLAIR MRI/DTI) and patients with the same imaging parameters.Each patient had IDH status obtained by gene sequencing (Training set:36 mutants and 40 wild types ;Test set : 25 mutants and 19 wild type). We have designed two kinds of multi-channel model including structure imaging model (Ts-net) and diffusion tensor imaging model (DTI-net) for compared, and finally merged them into the final mode(Tn-net). All images were co-registration to T1-CE MRI.The tumor and its vicinity were extracted by bounding-box that were manually drawn.Further validated by another radiologist to ensure that all tumor lesions were included.We standardized the 3D-patchs to 64x64x64 pixels to better serve as input for model.The Ts-net including T1-CE MRI,T2 MRI andT2-flair MRI sequence consisting of 3 channels as inputs. For DTI sequences,we used PANDA to process the main procedures including brain extraction eddy current correction and diffusion tensor and diffusion measure calculation.We calculated the 2 diffusion measure plot: fractional anisotropy (FA) and mean diffusion rate (MD) consisting of 2 channels as inputs. The final model contains all the sequences and consists of five channels as inputs.We used a kind of network called 3D-dencenet as the framework for training.We used a separate convolutional pooling layer for each channel and then attached the extracted feature vectors together before entering the fully connected layer.Each channel has a different weight.For all models, the inputs were 3D-patches, and the outputs were continuous values representing the likelihood of predicting IDH status. Internal validation was performed using tenfold and threefold cross-validation on training model to prevent model overfitting.To further verify the effectiveness of our model.we tested our training model on an independent test set from the TCIA database consisting of 44 patients.We preprocess the data according to the same procedure described in the data pretreatment section and perform receiver operating characteristics (ROC) studies to analyze the consistency between the classification generated by the model and the predicted results.Results
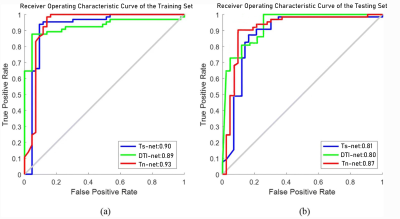
The performance measures averaged over all the folds are reported in Table 3, including accuracy (ACC), sensitivity (SEN), specificity (SPE), positive predictive rate (PPR), and negative predictive rate (NPR). The best classification accuracy of 90.17% can be achieved by combining the deep learning model with multiple modes. By contrast, using the structure image alone, we only achieved 82.35% accuracy. The test results on this independent data set are shown in figure 4. The performance of the test set is not much different from that of the training set. In the test set,0.87 of the area under the curve can be achieved by combining the deep learning model with multiple modes. This further proves the robustness of the proposed method.Discussion & Conclusion
We developed a multi-channel deep learning model based on 3d-Dencenet to predict IDH status of glioma patients and achieved good accuracy. More importantly, our proposed framework works well on small data sets and is validated on independent data sets to demonstrate the robustness of the proposed method. In addition, our proposed framework can fuse multi-modal information and combine more information from different imaging modes to determine the final classification to achieve better classification performance.Acknowledgements
NOReferences
1. Zhao Jing,Huang Yingqian,Song Yukun et al. Diagnostic accuracy and potential covariates for machine learning to identify IDH mutations in glioma patients: evidence from a meta-analysis.[J] .Eur Radiol, 2020, 30: 4664-4674.
2. Choi Yoon Seong,Bae Sohi,Chang Jong Hee et al. Fully automated hybrid approach to predict the IDH mutation status of gliomas via deep learning and radiomics.[J] .Neuro Oncol, 2021, 23: 304-313.
3. Eichinger Paul,Alberts Esther,Delbridge Claire et al. Diffusion tensor image features predict IDH genotype in newly diagnosed WHO grade II/III gliomas.[J] .Sci Rep, 2017, 7: 13396.
4. Abdel Razek Ahmed Abdel Khalek,Alksas Ahmed,Shehata Mohamed et al. Clinical applications of artificial intelligence and radiomics in neuro-oncology imaging.[J] .Insights Imaging, 2021, 12: 152.
5. Bangalore Yogananda Chandan Ganesh,Shah Bhavya R,Vejdani-Jahromi Maryam et al. A novel fully automated MRI-based deep-learning method for classification of IDH mutation status in brain gliomas.[J] .Neuro Oncol, 2020, 22: 402-411.
Figures