1412
Automatic segmentation of the hip bony structures on 3D Dixon MRI datasets using transfer learning from a neural network developed for the shoulder
Eros Montin1, Cem Murat Deniz1, Tatiane Cantarelli Rodrigues2, Soterios Gyftopoulos3, Richard Kijowski3, and Riccardo Lattanzi1,4
1Center for Advanced Imaging Innovation and Research (CAI2R) Department of Radiology, New York University Grossman School of Medicine, New York, NY, United States, 2Department of Radiology, Hospital do Coração (HCOR) and Teleimagem, São Paulo, Brazil, 3Department of radiology, New York University Grossman School of Medicine, New York, NY, United States, 4Vilcek Institute of Graduate Biomedical Sciences, New York University Grossman School of Medicine,, New York, NY, United States
1Center for Advanced Imaging Innovation and Research (CAI2R) Department of Radiology, New York University Grossman School of Medicine, New York, NY, United States, 2Department of Radiology, Hospital do Coração (HCOR) and Teleimagem, São Paulo, Brazil, 3Department of radiology, New York University Grossman School of Medicine, New York, NY, United States, 4Vilcek Institute of Graduate Biomedical Sciences, New York University Grossman School of Medicine,, New York, NY, United States
Synopsis
We describe a network for automatic segmentation of acetabulum and femur on 3D-Dixon MRI data. Given the limited number of labeled 3D hip datasets publicly available, our network was trained using transfer learning from a network previously developed for the segmentation of the shoulder bony structures. Using only 5 hip datasets for training, our network achieved segmentation dice of 0.719 and 0.92 for acetabulum and femur, respectively. More training data is needed to improve results for the acetabulum. We show that transfer learning can enable automatic segmentation of the hip bones using a limited number of labeled training data.
Introduction
Femoroacetabular impingement (FAI) is a pathological hip condition characterized by an abnormal shape of the bones of the hip joint, which results in contact between the acetabulum and femoral head–neck junction during hip motion 1. Computed Tomography (CT) has been used to create 3D patient-specific hip joint models to study FAI 2. However, such an approach results in potentially harmful ionizing radiations in the pelvis area. Furthermore, previous work required the manual segmentation of the region of interest, which is a time-consuming operation 2.It has been recently shown that MRI can be used not only for soft tissue (labrum and articular cartilage) assessment but also to reconstruct 3D models of the bony structures from a gradient-echo-based two-point Dixon sequence 3, which can eventually enable automatic bone segmentation.In fact, previous work using such pulse sequence demonstrated the reliability of a deep learning network for the automatic segmentation of the bones of the shoulders joint 4 (SHNET) and the proximal femur 5.
Based on the rationale that the shape of the hip and shoulder joints share many features, we propose to use transfer learning to develop a network (HNET) that is able to segment the bones of the hip joints using a customized version of the model of SHNET.
Materials and methods
DataThe Institutional Review Board approved this study and informed consent was obtained. Hip data of 7 female patients with an average age of 39 years old at the time of the exam were retrospectively retrieved. The MRI exam included a standardized Dixon acquisition with TR=10ms, 2 TE=2ms and 46ms, FlipAngle= 9, field-of-view 320mmx320mmx120mm, resolution 1x1x1mm centered on the patient's pelvis.
Methods
The SHNET’s architecture described in 3 was based on a 3D Convolutional Neural Network UNET composed of 4 layers, 64 filters, max-pool of 2x2x1, and size of 320x320x32 pixels (Figure 1).
A musculoskeletal radiologist (R.K) with more than 10 years of experience segmented the femur and the acetabulum on the 7 patients' Dixon3D datasets using ITK-snap 6.
HNET was trained using the Dixon images as input and the labeled images as output using a leave-one-out cross-validation approach with a learning rate of 1e-5, weighted cross entropy loss, 600 epochs and an early stopping condition on loss of 1e^-8.
Since each Dixon 3D dataset included both hips within the imaging field-of-view, while the SHNET had been trained on data that included only a single shoulder, we divided in half the hip volumes before training the HNET. In this way, the HNET network used 8 hips for training, 2 for testing and 4 for validation.
The training of HNET was repeated with random initialization of the network weights (RHNET), i.e., without transfer learning, to be used as a control performance.
Results
Figure 2 reports the results for the 4 validation data (4 hips from 2 subjects). For the femur, the average Dice coefficient was 0.719, 0.74 and 0.920 for the SHNET, RHNET and HNET (Jaccard 0.56,0.59, and 0.85, FP 0.39, 0.40, and 0.096, NP 0.11 and 0.304), respectively. For the acetabulum, the area Dice was 0.03, 0.558 and 0.67 SHNET, RHNET and HNET, respectively (Jaccard, 0.014, 0.387, and 0.50, FP 0.9853, 0.473 and 0.332 and FN 0.19, 0.406, and 0.33).Figures 3 and 4 report an example of the automatic segmentation on a validation subject's image. In yellow and blue the labels inferred by HNET, in red and green the manual segmentation
Discussion and conclusions
We presented a deep learning model for the segmentation of the bones of the hip joints based on the customization of a pre-existing model. This approach allowed us to train a neural network using a limited number of hip datasets, by exploiting the pre-existing knowledge inducted on shoulder data with SHNET. Note that the RHNET performed worse than HNET (Figure 2) implying that the performance achieved by the latter network depends on the initialization from the SHNET model.While the segmentation results for the femur were extremely promising, the relatively high rate of false negatives in the segmentation of the acetabulum suggests that a larger number of hip datasets might be needed to optimize the training.Our preliminary results proved that the proposed transfer learning approach could enable us to automatically segment the hip bones from 3D MRI data.
Our segmentation network could be incorporated into a fully automated pipeline to quantitatively evaluate FAI by using patient-specific 3D simulations of the hip range of motion and radiomic analysis on the bone shape and texture features.
Future work includes network refinement with additional segmented hip datasets and the release of HNET as part of Cloud MR, an open-source framework, currently in beta-testing, that uses cloud computing for the dissemination of MRI-related software tools 7-9.
Acknowledgements
HNET was developed through the Cloud MR project, which is supported in part by NIH R01 EB024536. This work was performed under the rubric of the Center for Advanced Imaging Innovation and Research (CAI2R, www.cai2r.net), a NIBIB Biomedical Technology Resource Center (NIH P41 EB017183).References
- Thomas, G. E. R., Palmer, A. J. R., Andrade, A. J., Pollard, T. C. B., Fary, C., Singh, P. J., O’Donnell, J., & Glyn-Jones, S. Diagnosis and management of femoroacetabular impingement. In British Journal of General Practice. 2013; (Vol. 63, Issue 612). https://doi.org/10.3399/bjgp13X669392
- Lerch, T. D., Degonda, C., Schmaranzer, F., Todorski, I., Cullmann-Bastian, J., Zheng, G., Siebenrock, K. A., & Tannast, M. Patient-Specific 3-D Magnetic Resonance Imaging–Based Dynamic Simulation of Hip Impingement and Range of Motion Can Replace 3-D Computed Tomography–Based Simulation for Patients With Femoroacetabular Impingement: Implications for Planning Open Hip Preservation Surgery and Hip Arthroscopy. American Journal of Sports Medicine.2019; 47(12), 2966–2977. https://doi.org/10.1177/0363546519869681
- Gyftopoulos, S., Yemin, A., Mulholland, T., Bloom, M., Storey, P., Geppert, C., & Recht, M. P. 3DMR osseous reconstructions of the shoulder using a gradient-echo based two-point Dixon reconstruction: A feasibility study. Skeletal Radiology. 2013; 42(3), 347–352. https://doi.org/10.1007/s00256-012-1489-z
- Cantarelli Rodrigues, T., Deniz, C. M., Alaia, E. F., Gorelik, N., Babb, J. S., Dublin, J., & Gyftopoulos, S. Three-dimensional MRI Bone Models of the Glenohumeral Joint Using Deep Learning: Evaluation of Normal Anatomy and Glenoid Bone Loss. Radiology: Artificial Intelligence. 2020; 2(5), e190116. https://doi.org/10.1148/ryai.2020190116
- Deniz, C. M., Xiang, S., Hallyburton, R. S., Welbeck, A., Babb, J. S., Honig, S., Cho, K., & Chang, G. Segmentation of the Proximal Femur from MR Images using Deep Convolutional Neural Networks. Scientific Reports. 2018; 8(1). https://doi.org/10.1038/s41598-018-34817-6
- Yushkevich, P. A., Piven, J., Cody Hazlett, H., Gimpel Smith, R., Ho, S., Gee, J. C., & Gerig, G. User-Guided 3D Active Contour Segmentation of Anatomical Structures: Significantly Improved Efficiency and Reliability. Neuroimage.2006; 31(3), 1116–1128.
- Montin E, Wiggins R, Block KT and Lattanzi R, MR Optimum – A web-based application for signal-to-noise ratio evaluation; 27th Scientific Meeting of the International Society for Magnetic Resonance in Medicine (ISMRM). Montreal (Canada), 11-16 May. 2019, p. 4617.
- Montin E, Carluccio G, Collins C and Lattanzi R, CAMRIE – Cloud-Accessible MRI Emulator; 28th Scientific Meeting of the International Society for Magnetic Resonance in Medicine (ISMRM). Virtual Conference, 08-14 August 2020, p. 1037
- Montin R, Carluccio G and Lattanzi R, A web-accessible tool for rapid analytical simulations of MR coils via cloud computing; 29th Scientific Meeting of the International Society for Magnetic Resonance in Medicine (ISMRM). Virtual Conference, 15-20 May 2021, p. 3756.
Figures
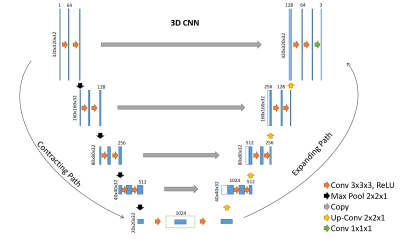
Architecture of the 3D CNNs used in this abstract. Blue rectangles represent feature maps with the size and the number of feature maps indicated on top of the bins. Different operations in the network are depicted by color-coded arrows. The architecture represented here contains 64 feature maps in the first and last layer of the network and 4 layers in the contracting/expanding paths.Dilated convolutions with multiple dilation rates are performed and concatenated at the center layer of 3D CNN dotted line in the center of the network
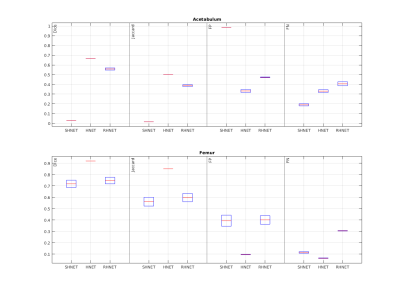
Segmentation results before (SHNET), after the transfer learning training session with the hip data (HNET), and using SHNET with random initialization of the weights (RHNET). Results for the Acetabulum (top) and Femur (bottom) are reported separately. In each subplot, an evaluation of the Dice and Jaccard coefficient along with the false positives (FP) and false-negative (FN) rates are reported for the three networks: SHNET, HNET and RHNET
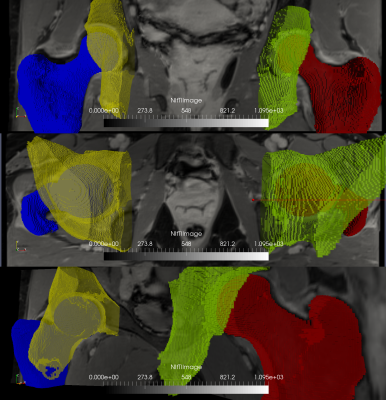
Automatic segmentation using HNET (yellow and blue) of the left hip is compared with the manual segmentation of the right hip (green and red), for a representative subject. The segmentations look similar in all three views (coronal, axial and semi-sagittal planes). Specifically, for this segmentation Dice was 0.67 and 0.92 for the acetabulum and femur, respectively (Jaccard: 0.50 and 0.85, FP: 0.32 and 0.09, FN: 0.35 and 0.066)
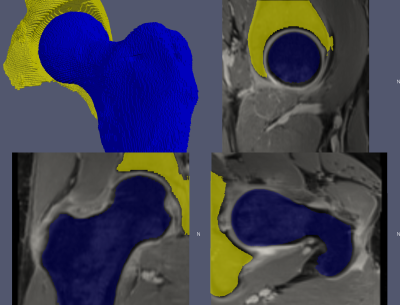
3D, sagittal, coronal, and axial views showing the outcome of the automatic hip segmentation using HNET for a representative hip overlayed to the input MR images. Note the error in the segmentation of the acetabulum in the sagittal view (top-right), due to similar image contrast between the bone and the tendon. While the segmentation for the femur was overall excellent, future network refinement with additional training datasets is needed to perfect the results for the acetabulum.
DOI: https://doi.org/10.58530/2022/1412