1410
Data-driven characterization of knee structures using non-negative matrix factorization of quantitative UTE Spiral VIBE MRI1imec-Vision Lab, Department of Physics, University of Antwerp, Wilrijk, Belgium, 2Department of Radiology, Antwerp University Hospital and University of Antwerp, Edegem, Belgium, 3Siemens Healthcare NV/SA, Beersel, Belgium
Synopsis
Model-driven methods for knee structure characterization, such as bi-exponential T2* mapping, may result in unreliable parameter estimation. In contrast, data-driven approaches, such as non-negative matrix factorization (NMF), allow to robustly identify multiple compartments. In this work, convexity-constrained NMF was used to decompose quantitative UTE MRI of three asymptomatic and one degenerative knees for structural characterization. Decomposition in four compartments led to minimal residual errors and low inter-subject differences in normalized mean compartment weights. Shifts in compartment weight distributions correlated to structural abnormalities, suggesting that the proposed approach may aid in the detection, grading and monitoring of internal knee derangements.
Introduction
The knee, one of the most complex joints in the human body, consists of multiple heterogenous structures that are individually characterizable by a mix of ultrashort (<1.0 ms), short (1–10 ms) and long (>10 ms) T2/T2* relaxation components.1 To capture the variety of relaxation values in knee structures, ultrashort echo time (UTE) sequences are commonly employed in combination with mono- and multi-exponential relaxation models.2,3 However, the use of multicompartment models often results in ill-conditioned problems and poses challenges to conventional fitting approaches. Consequently, data-driven methods have emerged as powerful alternatives for tissue characterization. Among them, non-negative matrix factorization (NMF) stands out by using non-negativity constraints which enables parts-based decomposition of data in an unsupervised fashion.4 NMF has previously been successfully applied to diffusion-weighted MRI for brain multi-tissue decomposition.5–7 The aim of this study is to explore NMF of quantitative UTE MRI for in vivo characterization of knee structures.Methods
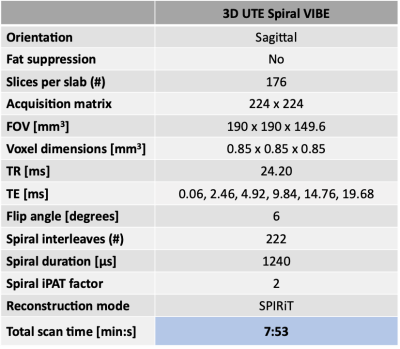
Two asymptomatic volunteers and one patient-volunteer (male, n=3; age, 30-38 years; BMI, 21.7-26.1 kg/m2) were scanned on a 3T MR scanner (Prisma Fit, Siemens Healthcare) with a 15-channel knee coil (QED). Datasets from 3 asymptomatic right knees and 1 degenerative left knee (20 years post-ACL reconstruction) were acquired with a commercially available anatomical 3D CAIPIRINHA SPACE TSE protocol8 and a UTE T2* mapping protocol based on an accelerated prototypical 3D UTE Spiral VIBE sequence9,10 (Table 1). T2*-weighted datasets were denoised using random matrix theory11 and corrected for Gibbs ringing artifacts.12 Rigid registration to the UTE data with lowest TE was performed in Elastix13 and signal intensity nonuniformity in T2*-weighted data was corrected with N4 bias field correction.14 The problem of identifying multiple compartments from a set of UTE T2*-weighted images was formulated as a convexity-constrained NMF problem15:$$ min_{{\bf W},{\bf H}}{\parallel{{\bf V}-{\bf VWH}} \parallel}_2^2 \ \ \ \ \ subject\ to \ \ \ \ {\bf W}=[w_{ij}] \geq 0,\ {\bf H}= [h_{ij}] \geq 0 $$
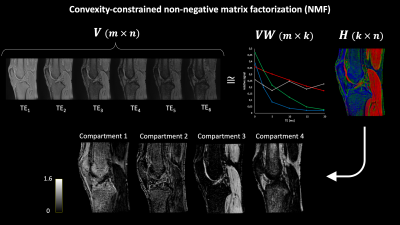
$$$ {\bf V} $$$ (m-by-n) represents the non-negative matrix of UTE T2*-weighted measurements, where each row corresponds to a different measurement (TE) and each column to a different voxel in the knee. $$$ {\bf W} $$$ (n-by-k) provides the weights of each voxel for each compartment. $$$ {\bf VW} $$$ (m-by-k) is defined as the basis matrix and $$$ {\bf H} $$$ (k-by-n) is the so-called weight matrix, where k is the rank of the factorization or the number of compartments to be retrieved from the data. NMF of a representative asymptomatic knee dataset is schematically depicted in Figure 1. Convergence of the iterative decomposition was defined based on the relative change in residual error between consecutive iterations. The optimal k was determined as the rank leading to the lowest residual error at convergence (k range: 2-6). The weight matrix of the degenerative knee was estimated based on the basis matrix of the contralateral asymptomatic knee. By means of the registered anatomical data, regions-of-interest (ROIs) were manually drawn in 9 knee structures and verified by an MSK radiologist. For each knee, mean NMF weights were calculated for every ROI and normalized to the sum of the compartments’ mean weights. The median of normalized mean weights was computed among asymptomatic knees for all ROIs. Mean normalized NMF weights of the degenerative knee were compared to the corresponding weights of the contralateral asymptomatic knee.
Results
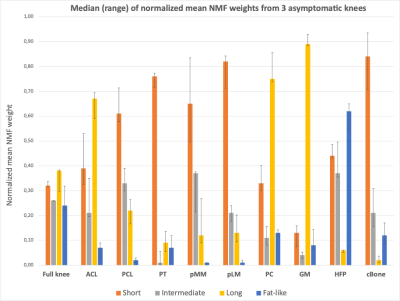
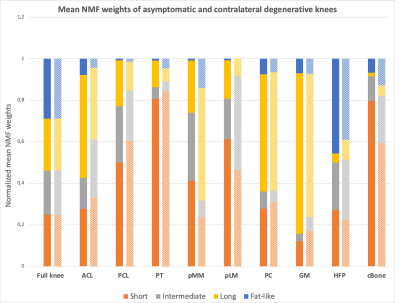
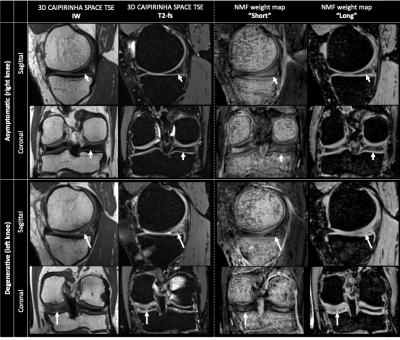
The optimal number of compartments for the NMF was found to be 4 as this resulted in the lowest residual error for all asymptomatic datasets. The shapes of the basis functions suggest that the input data can be decomposed into 3 components respectively displaying “short” (~3 ms), “intermediate” (~7 ms) and “long” (~27 ms) T2* decay, and 1 oscillating component related to fat (Figure 1). Medians of normalized mean NMF weights from 3 asymptomatic knees are shown in Figure 2. Knee structures with short mean transverse relaxation times, such as cancellous bone and patellar tendon, are predominantly composed of the “short” compartment, while long T2* structures such as patellar cartilage and muscle are largely comprised of the “long” compartment. Normalized mean NMF weights of one volunteer’s asymptomatic knee and contralateral degenerative knee are displayed in Figure 3. Considerable shifts in compartment weight distributions are observed for the menisci and bone of the degenerative knee.Discussion and Conclusion
NMF of quantitative UTE knee MRI resulted in the decomposition of knee structures in 4 compartments. Overall low inter-subject differences in normalized compartment weights indicate the potential of NMF to quantitatively characterize knee structures. The larger weight range in the posteromedial meniscus of asymptomatic knees resulted in the incidental finding of a low-grade intra-meniscal abnormality (Lotysch grade 1) (Figure 4).16 Moreover, comparative analysis of asymptomatic and degenerative knee data indicated that shifts in compartmental weight distributions correlate with observable structural abnormalities in e.g., the posteromedial meniscus (Figure 4).The proposed data-driven approach enables the characterization of knee structures based on quantitative UTE Spiral VIBE MRI, acquired in a clinically acceptable scan time, and thus holds promise as an instrumental aid for detection, grading and monitoring of structural knee abnormalities.
Acknowledgements
This project received funding from the UAntwerp SEP #44883.References
- Chang, E. Y., Du, J. & Chung, C. B. UTE Imaging in the Musculoskeletal System. J. Magn. Reson. Imaging 41, 870-883 (2015).
- Kijowski, R., Wilson, J. J. & Liu, F. Bicomponent ultrashort echo time T2* analysis for assessment of patients with patellar tendinopathy. J. Magn. Reson. Imaging 46, 1441–1447 (2017).
- Liu, J. et al. Single-and Bicomponent Analyses of T2 * Relaxation in Knee Tendon and Ligament by Using 3D Ultrashort Echo Time Cones (UTE Cones) Magnetic Resonance Imaging. Biomed Res. Int. (2019).
- Lee, D. D. & Seung, H. S. Learning the parts of objects by non-negative matrix factorization. Nature 401, 788–791 (1999).
- Jeurissen, B., Tournier, J.-D. & Sijbers, J. Tissue-type segmentation using non-negative matrix factorization of multi-shell diffusion-weighted MRI images. ISMRM 23rd Annu. Meet. Exhib. 20, 6755 (2015).
- Christiaens, D., Sunaert, S., Suetens, P. & Maes, F. Convexity-constrained and nonnegativity-constrained spherical factorization in diffusion-weighted imaging. Neuroimage 146, 507–517 (2017).
- Jillings, S. et al. Macro- And microstructural changes in cosmonauts’ brains after long-duration spaceflight. Sci. Adv. 6, (2020).
- Del Grande, F. et al. Fully Automated 10-Minute 3D CAIPIRINHA SPACE TSE MRI of the Knee in Adults A Multicenter, Multireader, Multifield-Strength Validation Study. Invest. Radiol. 53, 689–697 (2018).
- Qian, Y. & Boada, F. E. Acquisition-Weighted Stack of Spirals for Fast High-Resolution Three-Dimensional Ultra-short Echo Time MR Imaging. Magn. Reson. Med. 145, 135–145 (2008).
- Mugler, J. P. et al. Breath-Hold UTE Lung Imaging Using a Stack-of-Spirals Acquisition. Proc. Intl. Soc. Magn. Reson. Med. 23, (2015).
- Veraart, J. et al. Denoising of diffusion MRI using random matrix theory. Neuroimage 142, 394–406 (2016).
- Bautista, T., O’Muircheartaigh, J., Hajnal, J. V & Tournier, J.-D. Removal of Gibbs ringing artefacts for 3D acquisitions using subvoxel shifts. Proc. Intl. Soc. Magn. Reson. Med. 29, (2021).
- Klein, S., Staring, M., Murphy, K., Viergever, M. A. & Pluim, J. P. W. Elastix: A toolbox for intensity-based medical image registration. IEEE Trans. Med. Imaging 29, 196–205 (2010).
- Tustison, N. J., Avants, B. B., Cook, P. A. & Gee, J. C. N4ITK: Improved N3 bias correction. IEEE Trans. Med. Imaging 29, 708–711 (2010).
- Ding, C., Li, T. & Jordan, M. I. Convex and semi-nonnegative matrix factorizations. IEEE Trans. Pattern Anal. Mach. Intell. 32, 45–55 (2010).
- Crues, J. V, Mink, J., Levy, T. L., Lotysch, M. & Stoller, D. W. Meniscal Tears of the Knee: Accuracy of MR Imaging. Radiology 164, 445–448 (1987).
- Caldas de Almeida Araujo, E. Adaptation of Proof of Concepts Into Quantitative NMR Methods : Clinical Application for the Characterization of Alterations Observed in the Skeletal Muscle Tissue in Neuromuscular Disorders, Universite Paris Sud (2014).
Figures