1330
Workflow development for kidney segmentation using a U-net model using 11000 MRI data sets from the German National Cohort (NAKO/GNC) study1Magnetic Resonance Development and Applicatione Center, Medical Center, University of Freiburg, Faculty of Medicine, Freiburg, Germany, 2Medical Center, University of Freiburg, Faculty of Medicine, Freiburg, Germany
Synopsis
A processing pipeline for kidney segmentation using a hierarchical patch-based stack of U-nets was implemented and applied to abdominal MRI images of the German National Cohort study. The training data set included 300 cases and the final net was applied to the dataset of 11,207 MRIs. The compartments cortex, medulla and hilus could be segmented very robustly with the network. The relation of first parameters based on the segmentation withsex, age, weight, subject size and BMI are presented. This is an optimal starting point to identify more advanced biomarkers and their correlations, especially with kidney functional parameters.
Introduction
Chronic kidney disease (CKD) affects around 10% of adults worldwide and an estimated 13% to 17% in Germany. Imaging is a novel, promising approach to identify additional biomarkers of CKD and also of physiological kidney function[1-2]. Within a large, population-based cohort study, the German National Cohort study (NAKO/GNC), 30,000 participants underwent the MRI protocol described in [3]. The goal of our sub study is to identify relevant clinical, subclinical and biochemical correlates of the derived renal imaging biomarkers and to evaluate their performance in comparison to traditional markers of kidney function and damage in the GNC. In a first step, we aimed to develop a robust image processing pipeline for kidney segmentation and biomarker-characterization and apply it to abdominal MRI images from GNC study. The workflow will then be used to analyze the available baseline data as well as future follow up data.Methods
For the first 11,207 data sets, available 3D gradient echo and 2D haste images of the abdominal part of the data were imported into the imaging platform NORA (www.nora-imaging.org) for analysis. During this process, the data has been converted from DICOM to NIFTI format. The platform provides tools to manage, annotate and perform training of deep-learning convolutional neural networks (CNNs) on a large scale. For comparison, a subsample of 200 cases were initially segmented manually by two expert raters into the following compartments: cortex, hilus, medulla, and kidney cysts, similar to [4]. Initially already available, model-based algorithms for kidney segmentations (graph-cuts and others) have been applied. During this process it became clear that these approaches are not suitable to deal with the great inhomogeneity of image contrast, signal to noise, and the separability of kidneys and other tissue. Therefore, the focus has been set on deep-learning based approaches. A requirement analysis has been performed to design a convolutional-neural-network, and a U-Net model, configured using the “patchwork”-framework, developed in-house at the University Medical Center Freiburg (https://bitbucket.org/reisert/patchwork/wiki/Home), were applied. This hierarchical patch-based stack of U-nets was trained to segment these compartments automatically. Data was randomized, balanced, and mildly augmented on the fly. After a first training round, the model was iteratively improved by a loop of prediction, manual correction, and re-training in four rounds. The final model was trained on 300 cases and then applied to the dataset of 11,207 abdominal MRI. A manual quality control reading was performed to determine problematic cases and to select a valid dataset for statistical analysis. Based on the resulting segmentation, volumetric parameters (total kidney volume [TKV], sub-volumes of the compartments etc.) were calculated. The relative volumes of the compartments(normalized to TKV) cortex, hilus and medulla are related to sex, age, height, weight, and body-mass index (BMI) in univariate analyses.Results
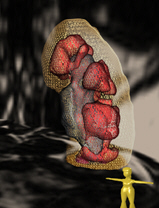
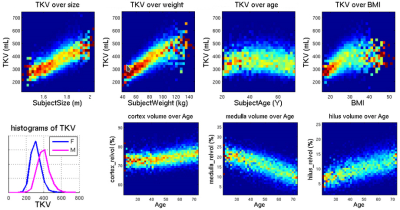
TKV and the three compartments cortex, medulla and hilus could be segmented very robustly with the trained network (Figure 1). Manual quality control revealed that segmentation quality was insufficient in 10% of the cases. This was predominantly due to initial imaging artifacts or bad image quality, and these cases were excluded from the statistical analysis. For the remaining data sets, the mean TKV was 416 mL for male and 330 mL for female participants. This is in line with previously reported values. As expected, there was a strong univariate association between height and weight of participants with TKV (Figure 2). Changes in relative cortex volume are more pronounced after age 45, similar to brain aging changes in gray and white matter.Discussion and conclusions
The developed framework allows for robust segmentation of kidneys in abdominal MRI data from a non-specific clinical routine protocol of a large cohort study. Basic parameters such as TKV and sub-compartment volumes of the kidney show correlations with participants’ height, weight, sex and age that are consistent with prior knowledge. Even though the used data was acquired on multiple sites within Germany, the scanner setup and used MR protocol was identical. Compared to most other multicentric studies, this leads to a high uniformity in data quality. Not only to better understand the requirements for the quality of the input data for such a workflow, it will be of interest to apply it also to data acquired on other scanners with similar but at least slightly variant protocols and therefore less uniform data. This is something every AI-based algorithm faces when applied not to study-based but real world clinical data. With this there is an optimal starting point to identify more advanced biomarkers (shape model, texture analysis, cyst count) and their correlates, especially kidney function parameters. This will help to better understand the prevalence of structural kidney variations in the general population and how established kidney disease markers relate to imaging-based biomarkers of kidney function and disease.Acknowledgements
This work funded by the Deutsche Forschungsgemeinschaft (DFG, German Research Foundation) – DFG KE 2513/1-1 / SPP 2177 within the DFG Priority Program SPP2177 „Radiomics: Nächste Generation der medizinischen Bildgebung“.References
[1] Eckardt KU, Coresh J, Devuyst O, et al. Evolving importance of kidney disease: from subspecialty to global health burden. Lancet 2013;382(9887):158-69. doi: 10.1016/S0140-6736(13)60439-0
[2] Aumann N, Baumeister SE, Rettig R, et al. Regional variation of chronic kidney disease in Germany: results from two population-based surveys. Kidney Blood Press Res 2015;40(3):231-43. doi: 10.1159/000368499
[3] Bamberg et. al. Whole-Body MR Imaging in the German National Cohort: Rationale, Design, and Technical Background. Radiology 2015, doi 10.1148/radiol.2015142272.
[4] Gloger et. al. Fully Automated Renal Tissue Volumetry in MR Volume Data Using Prior-Shape-Based Segmentation in Subject-Specific Probability Maps, IEEE Trans Biomed Eng 2015 Oct;62(10):2338-51. doi: 10.1109/TBME.2015.2425935.
Figures