1329
Radiomic biomarker extracted from PI-RADS 3 patients support more efficient and robust prostate cancer diagnosis: a multi-center study1the Collaborative Innovation Center for Internet Healthcare , School of Information Engineering, Zhengzhou University, Zhengzhou, China, 2Paul C. Lauterbur Research Center for Biomedical Imaging, Shenzhen Institute of Advanced Technology, Chinese Academy of Sciences, Shenzhen, China, 3Department of Urology, Renmin Hospital of Wuhan University, Wuhan, China, 4Department of Radiology, Guangdong Provincial People’s Hospital, Guangzhou, China
Synopsis
Prostate Imaging Reporting and Data System (PI-RADS) based on multi-parametric MRI classifies patients into 5 categories (PI-RADS 1-5) for routine clinical diagnosis guidance. However, there is no consensus on whether PI-RADS 3 patients should go through biopsies. Mining features from these hard samples (HS) is meaningful for physicians to achieve accurate diagnoses. Currently, the mining of HS biomarkers is insufficient, and the effectiveness and robustness of HS biomarkers for prostate cancer diagnosis have not been explored. In this study, biomarkers from different data distributions are constructed. Results show that HS biomarkers can achieve better performances in different data distributions.
Introduction
Prostate cancer is the most common type of solid organ malignancy in men worldwide[1]. Clinical studies have shown that prostate diseases present a differentiated clinical state from inert to highly aggressive[2]. Therefore, noninvasive and accurate diagnosis of clinically significant prostate cancer (csPCa) patients is very important, which can reduce excessive biopsy. Multi-parametric magnetic resonance imaging (mp-MRI) has already become an important diagnostic technique for prostate diseases. American College of Radiology and European Society of Urogenital Radiology proposed Prostate Imaging Reporting and Data System (PI-RADS) based on mp-MRI, which classified patients into 5 categories (PI-RADS 1-5) to aid in the diagnosis of csPCa according to the degree of malignancy with 1 being the lowest and 5 the highest [3-5]. However, severe controversies exist for the diagnosis of PI-RADS 3 patients regarding the necessity for them to go for biopsy [6,7]. Accordingly, these patients are treated as hard samples (HS), and more attention should be paid to these samples. Research in computer vision shows that putting more weight on the information extracted from difficult samples can help to improve the performance of the overall model [8-10]. Inspired by these successes, we believe that enhancing the information mining from mp-MRI of HS of prostate patients (PI-RADS 3 patients) can improve prostate cancer diagnostic performance. Nevertheless, most existing relevant studies perform prostate mp-MRI data mining from all collected samples without preference [11-13]. The biomarkers related to csPCa diagnosis contained in HS are not constructed properly, and the effectiveness and robustness of HS biomarkers have not been explored [14,15]. To this end, this study explores the effectiveness and robustness of radiomic biomarkers built with mp-MRI data of PI-RADS 3 patients for the diagnosis of csPCa. Experiments are conducted with data from three independent cohorts containing different distributions of samples, and results verify the effectiveness of the proposed method.Methodology
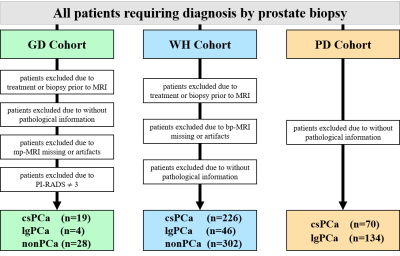
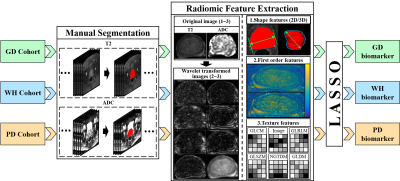
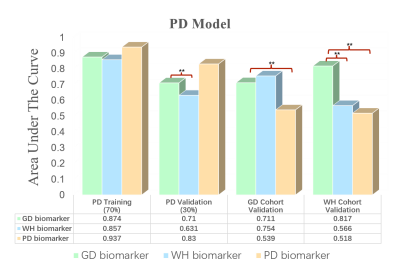
Detailed information of the experimental data utilized in this study is given in Figure 1. Among the three retrospective datasets, 204 patients in PD cohort are from the public data set of prostatex (Radboud University Medical Center), and the other two are from two medical centers in China, 574 patients in WH cohort (Wuhan University People's Hospital) and 51 patients in GD cohort (Guangdong Provincial People's Hospital). The PI-RADS evaluation of GD cohort and the segmentation of prostate in bp-MRI data of all patients were performed by two experienced doctors, and all results were examined by a senior expert. The proposed experimental procedure for obtaining the radiomic biomarkers is shown in Figure 2. A large number of quantitative radiomic features are obtained from the segmented prostate tissue. The least absolute shrinkage and selection operator (lasso) method with cross validation is used to perform feature selection and model construction. Three sets of radiomic biomarkers are obtained from the three cohorts with different prostate cancer patient distributions.Training and validation datasets in each cohort are partitioned with a ratio of 7: 3 based on patient visit time. The training dataset is used to construct the csPCa prediction model. In this study, three csPCa diagnostic models are constructed with the same training dataset using the three sets of radiomic biomarkers (extracted from the three cohorts). The diagnostic performance and robustness of the constructed models are validated on the validation datasets from all three cohorts. The performance of models is quantified by the area value under the curve. Delong tests are performed to check the significance of the performance difference between different models. The value of the information contained in the three sets of radiomic biomarkers is analyzed for prostate disease diagnosis.
Results and Discussion
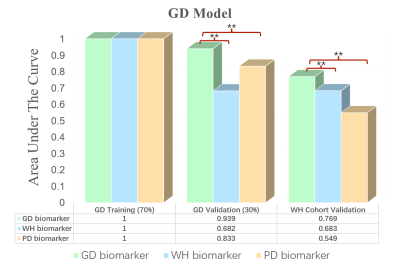
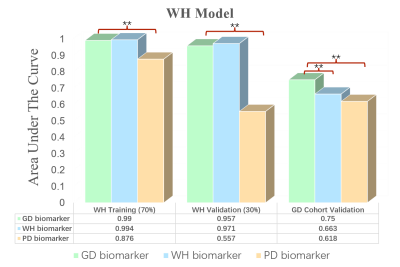
In total, 1576 features are extracted from bp-MRI of each patient, and three sets of radiomic biomarkers are obtained from the three cohorts (GD/WH/PD) by filtering these features. Here, the biomarkers of GD cohort are the biomarkers constructed from only HS (PI-RADS 3 patients), whereas for WH and PD, patients of other PI-RADS scores are also included in the biomarker construction process. Experimental results are shown in Figures 3-5. It can be observed that csPCa diagnostic models based on GD biomarkers have better diagnostic performances on all three cohorts, validating the effectiveness and robustness of HS biomarkers. We speculate that the radiomic biomarkers obtained from equivocal PI-RADS 3 patients can better capture the image representation differences between csPCa and inert prostate diseases. Therefore, we suggest that future csPCa diagnostic studies may pay more attention to image data mining of PI-RADS 3 patients.Conclusion
In this study, we found that the radiomic biomarkers obtained from PI-RADS 3 patients have better diagnostic value in the identification of csPCa. In future research on MRI-based diagnosis of csPCa, it is recommended to consider strengthening the data mining of PI-RADS 3 patients.Acknowledgements
This research was partly supported by theNational Natural Science Foundation of China (61871371, 81830056, 81801691,61671441), Youth Innovation Promotion Association Program of Chinese Academy ofSciences (2019351), Collaborative Innovation Major Project of Zhengzhou (20XTZX06013).References
[1] Fitzmaurice C, Allen C, Barber R M, et al.Global, regional, and national cancer incidence, mortality, years of life lost, years lived with disability, and disability-adjusted life-years for 32 cancer groups, 1990 to 2015: a systematic analysis for the global burden of disease study[J].JAMA oncology,2017, 3 (4): 524-548.
[2] Siegel R L, Miller K D, Jemal A.Cancer statistics, 2020[J].Ca-a Cancer Journal for Clinicians,2020, 70 (1): 7-30.
[3] Barentsz J O, Richenberg J, Clements R, et al.ESUR prostate MR guidelines 2012[J].Eur Radiol,2012, 22 (4): 746-57.
[4] Weinreb J C, Barentsz J O, Choyke P L, et al.PI-RADS prostate imaging–reporting and data system: 2015, version 2[J].European urology,2016, 69 (1): 16-40.
[5] Turkbey B, Rosenkrantz A B, Haider M A, et al.Prostate Imaging Reporting and Data System Version 2.1: 2019 Update of Prostate Imaging Reporting and Data System Version 2[J].Eur Urol,2019, 76 (3): 340-351.
[6] Wadera A, Alabousi M, Pozdnyakov A, et al.Impact of PI-RADS Category 3 lesions on the diagnostic accuracy of MRI for detecting prostate cancer and the prevalence of prostate cancer within each PI-RADS category: A systematic review and meta-analysis[J].Br J Radiol,2021, 94 (1118): 20191050.
[7] Schoots I G.MRI in early prostate cancer detection: how to manage indeterminate or equivocal PI-RADS 3 lesions?[J].Translational andrology and urology,2018, 7 (1): 70.
[8] Lin T Y, Goyal P, Girshick R, et al.Focal Loss for Dense Object Detection[J].IEEE Transactions on Pattern Analysis & Machine Intelligence,2017, PP (99): 2999-3007.
[9] Shrivastava A, Gupta A, Girshick R. Training Region-based Object Detectors with Online Hard Example Mining[C].IEEE Conference on Computer Vision & Pattern Recognition,2016: 761-769.
[10] Sheng H, Zheng Y, Ke W, et al.Mining Hard Samples Globally and Efficiently for Person Re-identification[J].IEEE Internet of Things Journal,2020, PP (99): 1-1.
[11] Yuan Y, Qin W, Buyyounouski M, et al.Prostate cancer classification with multiparametric MRI transfer learning model[J].Medical physics,2019, 46 (2): 756-765.
[12] Winkel D J, Tong A, Lou B, et al.A Novel Deep Learning Based Computer-Aided Diagnosis System Improves the Accuracy and Efficiency of Radiologists in Reading Biparametric Magnetic Resonance Images of the Prostate: Results of a Multireader, Multicase Study[J].Invest Radiol,2021, 56 (10): 605-613.
[13] Zhang Y, Chen W, Yue X, et al.Development of a Novel, Multi-Parametric, MRI-Based Radiomic Nomogram for Differentiating Between Clinically Significant and Insignificant Prostate Cancer[J].Front Oncol,2020, 10: 888.
[14] Hectors S J, Chen C, Chen J, et al.Magnetic Resonance Imaging Radiomics-Based Machine Learning Prediction of Clinically Significant Prostate Cancer in Equivocal PI-RADS 3 Lesions[J].J Magn Reson Imaging,2021, 54 (5): 1466-1473.
[15] Görtz M, Radtke J P, Hatiboglu G, et al.The Value of Prostate-specific Antigen Density for Prostate Imaging-Reporting and Data System 3 Lesions on Multiparametric Magnetic Resonance Imaging: A Strategy to Avoid Unnecessary Prostate Biopsies[J].Eur Urol Focus,2021, 7 (2): 325-331.
Figures