1328
Developing a Craniofacial Soft Tissue Anthropomorphic Database: Applying Diffeomorphic Template Building Methods to MRI1Children's Hospital of Philadelphia, Philadelphia, PA, United States, 2Perelman School of Medicine at the University of Pennsylvania, Philadelphia, PA, United States
Synopsis
Craniofacial surgery requires reconciliation of several factors for deformity reconstruction and aesthetic enhancement. Here, we present data in developing a racially and ethnically sensitive anthropomorphic database to provide plastic and craniofacial surgeons with a set of “normal” anatomic measurements to optimize aesthetic and reconstructive outcomes. Images were used to construct composite (template) images with Advanced Normalization Tools (ANTs) and Greedy Tools. Composite templates demonstrated age-appropriate anatomic measurements as proof of concept. Application of diffeomorphic algorithms via ANTs to MRI is effective in developing composite templates representing “normal” soft tissue anatomy, which may aid in objectively quantifying anatomic abnormality.
Introduction
Craniofacial surgery requires reconciliation of cephalometric, anthropomorphic, cultural, and experience-based factors for deformity reconstruction and aesthetic enhancement. In the pediatric population, assessing short- and long-term postoperative outcomes may prove particularly challenging due to rapid craniofacial growth, often anticipated with surgical “overcorrection.” In this study, the authors present data in developing a racially and ethnically sensitive anthropomorphic database to provide plastic and craniofacial surgeons with a set of “normal” anatomic measurements to optimize aesthetic and reconstructive outcomes.Methods
A list of head MRIs from 2008 to 2021 at the authors’ institution was retrospectively reviewed. Patients were included if they received a 3T head MRI for indications without craniofacial implication and had no preexisting pathology altering craniofacial and soft tissue structures. T1-weighted images were obtained at 3T (TIM Trio; Siemens Medical Solutions, Erlangen, Germany) at 1 mm3 isotropic resolution. Each of the MRI images were thresholded and manually corrected by an expert using ITK-SNAP.1 Images were used to construct template (composite) images with Advanced Normalization Tools (ANTs)2 and Greedy Tools.3 The algorithms were based on both symmetric diffeomorphic image registration with cross-correlation and a diffeomorphic image averaging approach, alternating between averaging all image intensity and registering all images to the intensity average (Figure 1, upper).4 The template creation approach was as follows: I0 was initialized to arithmetic mean of J1, J2, … JN (Figure 1, upper). For m = 0 … M, where “m” is the loop iteration, each image Ji was registered to Im. Each image Ji was deformed to obtain Ĵi, and the new mean template “Im+1 ” was constructed using Ĵim. Template MRIs were thresholded to generate binary segmentations and used to generate quantitative anatomic renderings in Materalise Mimics v23 (Materalise, Ghent, Belgium)5 (Figure 1, lower right).Results
Sixteen templates were generated: four each from three-, four-, seven-, and eight-year-olds (two male, two female, two black, and two white) from ten MRI sequences, each. The average head circumferences were on average 2.3 cm larger in the four-year-old templates (51.8 cm) compared to the three-year-old-templates (49.5 cm), and 1.0 cm larger in the eight-year-old templates (53.9 cm), compared to the seven-year-old templates (52.9 cm). The average measured lateral canthus to lateral canthus distances increased from 81.2 mm to 86.6 mm in the three- to four-year-old templates and 88.1 mm to 90.7 mm in the seven- to eight-year-old templates. Nasion to nasal tip distance was on average 1.1 mm longer in the four-year-old templates (24.1 mm) compared to the three-year-old templates (23.0 mm) and 4.4 mm longer in the eight-year-old templates (31.4 mm) compared to the seven-year-old templates (26.9 mm). Average maximal ear height was on average 3.1 mm greater in the four-year-old templates (55.4 mm) compared to the three-year-old templates (52.3 mm) and 0.8 mm greater in the eight-year-old templates (61.7 mm) compared to the seven-year-old templates (60.9 mm).Conclusions
Application of diffeomorphic algorithms via ANTs to MRI is effective in creating composite templates to represent “normal” craniofacial and soft tissue anatomy, which may aid in assessing short- and long-term postoperative outcomes. Further work is needed to optimize accuracy and precision of composites with increased power from additional MRI images for each demographic of interest. Future research will focus on development of mathematical tools to characterize anatomic normality, generation of indices to grade preoperative severity, and quantification of postoperative surgical results to reduce subjectivity bias.Acknowledgements
This research was supported by the Division of Plastic & Reconstructive Surgery at the Children’s Hospital of Philadelphia and the Penn Image Computing and Science Laboratory, Department of Radiology at the University of Pennsylvania Perelman School of Medicine.References
1. Yushkevich, P. A., Yang, G., Gerig, G. ITK-SNAP: An interactive tool for semi-automatic segmentation of multi-modality biomedical images. Conference proceedings: Annual International Conference of the IEEE Engineering in Medicine and Biology Society IEEE Engineering in Medicine and Biology Society Annual Conference 2016;2016:3342-3345.
2. Avants, Brian B., Nick Tustison, and Gang Song. "Advanced normalization tools (ANTS)." Insight j 2.365 (2009): 1-35.
3. Venet, L., Pati, S., Yushkevich, P., & Bakas, S. (2019). Accurate and robust alignment of variable-stained histologic images using a general-purpose greedy diffeomorphic registration tool. arXiv preprint arXiv:1904.11929.
4. Yushkevich, P.A., Pluta, J., Wang, H., Wisse, L.E., Das, S. and Wolk, D., 2016. Fast Automatic Segmentation of Hippocampal Subfields and Medial Temporal Lobe Subregions in 3 Tesla and 7 Tesla MRI. Alzheimer's & Dementia: The Journal of the Alzheimer's Association, 12(7), pp.P126-P127.
5. https://www.materialise.com/en/medical/mimics-innovation-suite
Figures
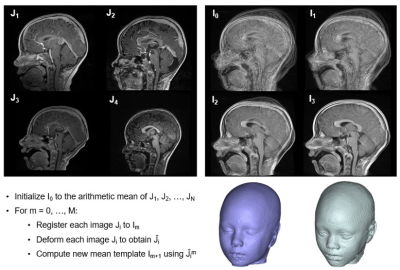
Figure 1. Upper left: sample images of subjects used to create a template; upper right: iterative template creation steps; lower left: brief overview of algorithmic approach used to create template images; lower right: sample resultant template images of seven-year-old white female (left) and eight-year-old black female (right).