1285
An iterative algorithm for resolving high-resolution 23Na Multi-Quantum Coherences MRI from prior 1H constraints1Computer Assisted Clinical Medicine, Heidelberg University, Mannheim, Germany, 2CRMBM, Aix Marseille University, CNRS, Marseille, France
Synopsis
Sodium (23Na) MRI offers great potentials to be a clinical marker for disease states. Besides the single quantum signal, the triple quantum signal of 23Na could provide novel and complementary information. 3D volumetric 23Na multi-quantum coherences imaging, however, suffers from poor signal-to-noise ratios, which limit spatial resolution. Finer structures such as tissue boundaries are therefore barely observable. In addition, partial volume effects become more severe and diminish image quality as well as diagnostic applicability. The purpose of this work is to develop a reconstruction framework that addresses these shortcomings of 23Na multi-quantum coherences imaging by utilizing 1H prior constraints.
Introduction
23Na nuclei exhibit a quadrupolar moment and under certain conditions, one can observe besides the conventional single quantum coherences, also triple quantum (TQ) coherences. The TQ signal is very sensitive to its sodium environment1 and could therefore provide additional information about the tissue and the state of the disease2. Thus, further developments are warranted in 23Na multi-quantum coherences (MQC) imaging. The 23Na signal's intensity holds correlation with the 23Na in-vivo concentration3 but suffers from poor SNR. In parallel, 1H MRI offers superior SNR and fine morphological images with resolutions in the sub millimeter range but lacks access to tissue homeostasis information. Thus, we hypothesized that 1H information can be utilized to enhance known tissue boundaries in 23Na MRI4 to improve 23Na MQC imaging.Methods
Imaging was performed on 3T (phantom) and 7T (in-vivo) MRI (Trio and Magnetom, Siemens) with a 1Tx/Rx dual-tuned 1H/23Na head coil. Prior to the MQC scans, B0 shimming was repeated with a 1H-based vendor provided 3D shimming routine until convergence. A home-built B1+ calibration sequence was employed to calibrate global 90° RF pulses. The optimal MQC evolution time τ was determined from a global TQTPPI spectroscopic prescan, followed by an offline fit.Phantom: FoV 276x276x200mm3, matrix size 32x32x10 and TR=140ms resulting in a total of 62 minutes.
In-vivo: Three healthy volunteers were scanned to demonstrate in-vivo feasibility. 20 TEs, with TE1=0.96ms (asymmetric echo, ΔTE=3ms), TR=390ms, phase increment Δφ=30° (12-steps phase cycle), matrix size 64x64, slice thickness 20mm, FOV=224mm and a bandwidth of 330Hz/Px, resulting in a total acquisition time of TA=50min. Anatomical information of the brain was obtained via multi-slice 2D 1H TSE sequence with 1.2x1.2x3mm3, TE= 66ms, TR= 13.6s, BW= 224 Hz/px and TA=4:19 min.
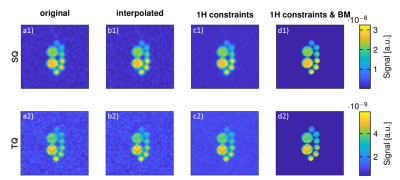
TQ and SQ images were reconstructed prior to data processing as described previously5. To match the resolution of the 1H image and to ensure better image registration, 23Na MQC data was interpolated to the same size as the 1H image. Finally, interpolated 23Na data were processed to limit partial volume effects with constraints from the 1H prior first order total variation algorithm:
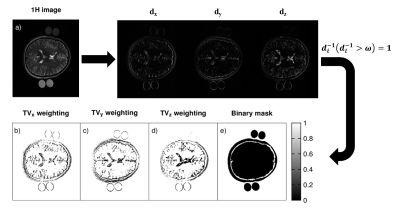
$$\underset{u}{{min}} \,\, \lambda_{xyz} ||\nabla_x u, \nabla_y u, \nabla_z u||_2 + \lambda_{BM} ||BM*u||_2 \ \, \,s.t. \, \, ||\varPhi_F (u) - f||^2_2 < \sigma^2 $$
With $$$u$$$ being the targeted improved interpolated image, $$$\varPhi_{F}$$$ indicating the partial Fourier transform and $$$f$$$ representing the measured data in frequency domain and $$$\sigma$$$ being the signal variance noise. 1H prior information is used to detect sharp edges by calculating the first derivative along $$$x,y,z$$$ of the high resolution 1H image. The derivatives are inverted and can be regarded as threshold maps to locally enhance edges and support TV denoising. The amount of included prior information can be controlled by setting a threshold, $$$ω$$$. $$$\lambda_{BM}||BM*u||_2$$$ represents another regularization term that penalizes the area outside of the region of interest, e.g. air in order to further reduce partial volume effects. $$$\lambda_{xyz}$$$ and $$$\lambda_{BM}$$$ are weighting parameters for the edge enhancing and the outer region penalizing regularization term, respectively. The Split-Bregman was utilized to solve the optimization problem stated in equation 1 with the following empirically determined weighting factors $$$\lambda_{xyz}=1$$$ and $$$\lambda_{BM}=0.4$$$. GPU computing was utilized resulting in a total reconstruction time of 140 seconds. To evaluate sharpness of an image, the sum of the absolute Wavelet coefficients (WAVS)6 was used. All calculations were performed in MATLAB (MATLAB R2020a, MathWorks, Natick, MA).
Results
Figure 1 demonstrates the 1H TSE image and the computed thresholding maps to locally enhance and support TV denoising.It is visible in Figure 2 that the proposed iterative reconstruction with 1H prior constraints reduces partial volume effects.
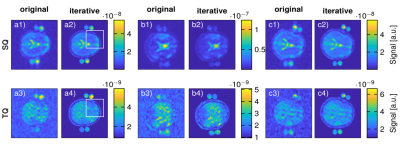
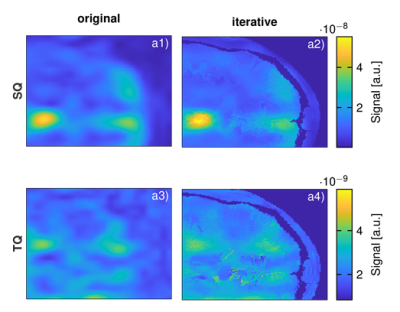
Figure 3 shows the comparison of regular 23Na MQC and the proposed iterative reconstruction in-vivo. A clear distinction between the brain, the skull and the outer skin for all three volunteers is possible. Thus, the iterative reconstruction provides sharper images when compared to the standard reconstruction, WAVSSQ=(1.07±0.37)*10-4, WAVSTQ = (5.59±1.54)*10-4 , WAVSSQiter=(1.12±0.26)*10-3, WAVSTQiter = (2.9±0.59)*10-3.
Discussion
While the proposed framework enabled to reach virtual high resolution sodium MQC imaging, final images might locally appear grainy from the inherent noise in 1H images. For instance in the mask, the cerebrospinal fluid exhibited speckles that could result in noise addition in the final 23Na image. This could be alleviated with filtered 1H images and/or the registration of all images in an atlas space.Further processing could also include anatomical information from 1H T1-weighted images or maps that identify specific regions such as white and gray matter to constrain their relative sodium intensity.
Fiege et. al.7 were not able to evaluate TQ information in large brain tumours due to limited MQC image resolution. The proposed iterative framework could complement TQ information as tumors, or other targeted lesion, could be well delineated in 1H images.
Conclusion
We developed an iterative reconstruction algorithm that improves 23Na MQC image quality by utilizing prior 1H MR image information to preserve known tissue boundaries in Single and Triple Quantum sodium images.Acknowledgements
No acknowledgement found.References
1. LaVerde, G., Nemoto, E., Jungreis, C., Tanase, C. and Boada, F. (2007), Serial triple quantum sodium MRI during non-human primate focal brain ischemia. Magn. Reson. Med., 57: 201-205. https://doi.org/10.1002/mrm.21087.
2. Hoesl MAU, Kleimaier D, Hu R, et al. 23Na Triple-quantum signal of in vitro human liver cells, liposomes, and nanoparticles: cell vi- ability assessment vs. separation of intra- and extracellular signal. J Magn Reson Imaging. 2019;50:435-444.
3. Huhn K, Engelhorn T, Linker RA, Nagel AM. Potential of Sodium MRI as a Biomarker for Neurodegeneration and Neuroinflammation in Multiple Sclerosis. Front Neurol. 2019 Feb 11;10:84. doi: 10.3389/fneur.2019.00084
4. Gnahm, C., Nagel, A. ,Anatomically weighted second-order total variation reconstruction of 23Na MRI using prior information from 1H MRI,NeuroImage,Volume 105,2015,Pages 452-461,ISSN 1053-8119.
5. Hoesl, M.A.U., L.R. Schad, and S. Rapacchi, Efficient 23Na triple-quantum signal imaging on clinical scanners: Cartesian imaging of single and triple-quantum 23Na (CRISTINA). Magnetic Resonance in Medicine, 2020. 84(5): p. 2412-2428.
6. Jain A., et al., Robust technique of depth focus measure in Wavelet domain, IOSR Journal of VLSI and Signal Processing (IOSR-JVSP) Volume 6, Issue 5, Ver. II (Sep. - Oct. 2016), PP 07-14 e-ISSN: 2319 – 4200, p-ISSN No. : 2319 – 4197.
7. Fiege, D.P., Romanzetti, S., Mirkes, C.C., Brenner, D. and Shah, N.J. (2013), Simultaneous single-quantum and triple-quantum-filtered MRI of 23Na (SISTINA). Magn Reson Med, 69: 1691-1696. https://doi.org/10.1002/mrm.24417 .
Figures