1263
Improving T1 mapping with a golden-angle radial MOLLI and a model-based regularized reconstruction SALSA-EPG
Andreia C Freitas1, Andreia S Gaspar1, Nuno A Silva2, José M Bioucas-Dias3, and Rita G Nunes1
1ISR Lisboa/LARSyS and Department of Bioengineering, Instituto Superior Técnico - Universidade de Lisboa, Lisbon, Portugal, 2Hospital da Luz Learning Health, Luz Saúde, Lisbon, Portugal, 3Instituto de Telecomunicações, Instituto Superior Técnico - Universidade de Lisboa, Lisbon, Portugal
1ISR Lisboa/LARSyS and Department of Bioengineering, Instituto Superior Técnico - Universidade de Lisboa, Lisbon, Portugal, 2Hospital da Luz Learning Health, Luz Saúde, Lisbon, Portugal, 3Instituto de Telecomunicações, Instituto Superior Técnico - Universidade de Lisboa, Lisbon, Portugal
Synopsis
T1 mapping provides valuable information regarding cardiovascular pathologies. Clinical approach consists of using a Cartesian MOLLI sequence and fitting a 3-parameter model. Although easy and straightforward, this method can require long breath-holds and does not explicitly include other factors affecting T1 estimation. We propose coupling an accelerated golden-angle radial MOLLI with a model-based regularized reconstruction (SALSA). The proposed method was tested in phantom and in vivo data at 1.5T. Improved T1 precision and good accuracy was found for the phantom data. Lower T1 was estimated with SALSA compared to the commercial sequence in vivo, future work will address this.
Introduction
Quantitative MRI provides valuable information regarding cardiovascular pathologies as myocardial T1 changes may reflect infarction, diffuse fibrosis or inflammation1. The clinical approach consists of acquiring multiple T1-weighted images with a Cartesian MOLLI (MOdified Look-Locker) sequence and fitting a 3-parameter model: S(TI)=A–B.exp(-TI/T1*); where T1~T1*(B/A-1). However, other factors that affect the magnetization state and lead to T1 bias are not explicitly taken into account (e.g. effective flip angle, heart rate variability and others). Another issue of conventional cartesian MOLLI is the lengthy breath-hold required. To address this, we propose coupling an accelerated golden-angle (GA) radial MOLLI with a model-based regularized reconstruction (SALSA).Methods
An in-house open-source radial GA (with alternating spoke direction) trajectory with a MOLLI 5(3)3 scheme was considered (flip angle = 35º, 2 inversions with inversion time (TI) = 104 and 186ms, TR=3.03ms, FOV=200mm2 and trigger delay=400ms). The pulse sequence was implemented with Pulseq2 and data acquired on a Siemens Aera 1.5T scanner. Preliminary testing was performed on the ISMRM/NIST T1/T2 phantom and in vivo on skeletal leg muscle with a simulated ECG (50 beats per minute). Radial GA data was acquired at different acceleration rates from fully sampled (R =1) to undersampled (R = 4). T1-weighted radial images were then reconstructed offline with NUFFT3 and SALSA4. Zeroth-order phase corrections were applied to k-space before reconstruction; i.e. the phase of k-space centre in each radial spoke was calculated and subtracted to all other spoke readout points. An Extended Phase Graph (EPG) dictionary was computed considering the acquisition parameters and a T1 range of [5:5:2400]ms and T2 of [10:10:1000]ms. MR image reconstruction can be described as y=Bx+n, where y is the acquired noisy k-space data, x the reconstructed image and B is the direct operator (here including the Fourier transform). SALSA solves the constrained optimization problem min 1/2 ||y-M(EXF)||+||d X W|| where M is the mask which defines the undersampling of the acquired k-space y, E is the compressed dictionary computed with EPG, F is the Fourier operator (which in the non-Cartesian case corresponds to NUFFT), W is the 2D discrete wavelet operator of the rows of X (matrix of coefficients combining the compressed dictionary components) and d is a diagonal matrix controlling the weight of W. This can then be solved by using an alternating direction method (ADMM). T1 maps were obtained by 3-parameter fit to the accelerated T1-w images, and by matching the NUFFT and SALSA T1w-images to the dictionary. To serve as a comparison, T1 maps were also acquired with the commercial Cartesian MOLLI sequence (TI = 231/331ms) for both phantom and in vivo. A schematic representation of the methodology is seen in Fig 1.Results
Reduction of aliasing artefacts (accelerated radial R = 4) can be seen in T1-w images reconstructed with SALSA (for phantom studies, λ=1e-5 was chosen) compared to standard NUFFT (Fig 2). Mean T1 values measured in each phantom vial were compared to the T1 ground truth from the NIST phantom manual (Fig 2 j - dark blue bar) and the commercial Cartesian MOLLI sequence (light blue bar). Accelerated radial (allowing to reduce breath-hold time compared to Cartesian MOLLI) demonstrated comparable T1 measurements to the commercial sequence for vials 2-4 (corresponding to the range of more relevant T1 = 1489 to 780ms). However, an underestimation was observed for the dictionary methods (NUFFT and SALSA) for vial 1 (T1=2033 ms). SALSA also resulted in less T1 spatial dispersion within each vial (lower variation coefficient - Fig 2 k) compared to NUFFT. T1 maps were also computed for leg muscle data sets (Fig 3), where a λ= 3e-6 was chosen. Similarly to phantom results, reduction of the aliasing artefacts can be seen in the accelerated radial SALSA T1-w images (Fig 3 f) compared to NUFFT. As expected, the SALSA method leads to improved precision compared to the NUFFT T1 maps (lower variation coefficient - Fig 3 k). However, mean T1 values of the SALSA maps appear to be slightly lower compared to the commercial MOLLI sequence.Discussion and conclusions
The proposed radial GA MOLLI with SALSA-EPG appears to be a promising method to obtain fast open-source T1 mapping. Accelerations up to 4-fold were tested in the NIST phantom and leg muscle, demonstrating the potential of the proposed method for reducing the overall breath-hold duration of standard MOLLI. Good T1 accuracy and precision was obtained with the SALSA method compared to the ground truth and the commercial sequence for the T1 range most relevant for a later cardiac application (1489-780ms). However, estimated T1 values in vivo were lower with the proposed method than the commercial MOLLI. Potential reasons for this could be the choice of TI used in the radial sequences (shorter than the commercial), and differences between off-line and in-line estimation procedures (the MOLLI Cartesian map was obtained using Siemens reconstruction and estimation pipeline). Future work will attempt to improve the accuracy of the SALSA-EPG by optimizing the chosen TI, including coil sensitivity information in the SALSA optimization and correcting for first-order gradient-induced phase errors.Acknowledgements
Fundação para a Ciência e a Tecnologia (SFRH/BD/120006/2016, PTDC/EMD-EMD/29686/2017) e Programa Operacional Regional de Lisboa 2020 (LISBOA-01-0145-FEDER-029686)References
[1]Taylor, JACC: Card Img(2016)9:67-81 [2] Layton, MRM (2017) 77:1544-1552 [3]Fessler, JMR(2007)188:191-195 [4]Afonso, IEEE Trans Img Process(2010)19:2345-56Figures
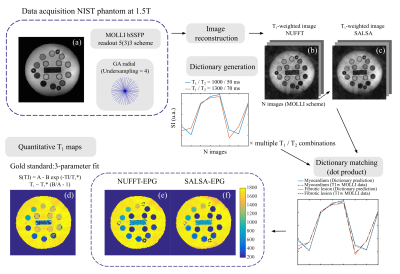
Schematic of T1 mapping computing environment. (a) Phantom with 14 vials representing a T1 range (22-2033ms). Open-source radial GA trajectory implemented with a 5(3)3 MOLLI scheme. Accelerated data acquired and reconstructed offline using (b) NUFFT and (c) SALSA. EPG dictionary computed considering the sequence parameters and a range of T1/T2 values. T1-w images were matched to the dictionary and T1 maps computed (e-f). T1 mapping with the 3-parameter fitting method was also computed (d).
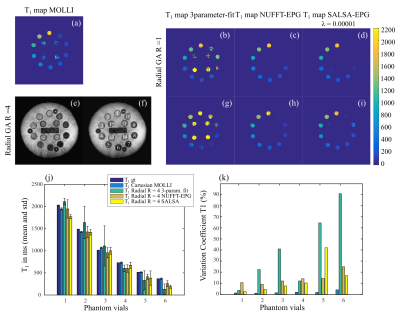
Phantom results. T1 maps computed with (a) the commercial MOLLI, (b-d) the radial GA fully sampled data and accelerated 4-fold radial data (g-i) and the 3 described methods. (e-f) Example of T1-w images reconstructed from accelerated radial data with NUFFT and SALSA. (j) Mean and standard deviation T1 compared to ground truth values. Only vials with the more relevant T1 values for the in vivo case were included. (k) Variation coefficient (standard deviation divided by the mean T1).
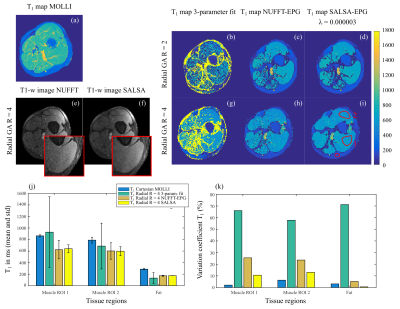
In vivo leg muscle T1 mapping results. (a) T1 map obtained with the commercial MOLLI sequence. (b-d) T1 maps were obtained with the accelerated radial GA R = 2 (200 spokes), (g-i) radial R = 4 data (100 spokes) and the three described methods. (e-f) Example of T1-w images reconstructed from accelerated radial GA data with NUFFT and SALSA. (j) Mean and standard deviation T1 for each method, compared to the commercial MOLLI. (k) Variation coefficient (standard deviation divided by the mean T1).
DOI: https://doi.org/10.58530/2022/1263