1165
Intrinsic reproducibility issues in deep learning-based MR reconstruction1Department of Electrical and Computer Engineering, Seoul National University, Seoul, Korea, Republic of, 2AIRS Medical Inc., Seoul, Korea, Republic of
Synopsis
Deep learning requires a large number of parameter settings, which can be prone to reproducibility issues, undermining the reliability and validity of the outcomes. In this study, we list the common sources that may induce the reproducibility issues in deep learning-based MR reconstruction. The effect size of each source on the network performance was investigated. From the results of this study, we recommend to share a trained network.
Introduction
Recently, a number of areas including parallel imaging1 and quantitative susceptibility mapping2 have utilized deep learning-based MR reconstruction to improve performances. Despite the gains, however, questions have risen regarding the generalization of the network performance for the inputs of different scan parameters3 as well as the reliability of the reconstruction for small perturbation in input images.4 Another venue to explore network characteristics is reproducibility. In deep reinforcement learning, for example, studies have shown that networks suffer from low reproducibility due to sources like initialization seeds,5,6 limiting validation of the results. More generally, numerical operations in neural networks have been suggested to have reproducibility issues.7In this study, we tested a list of potential sources that may induce “intrinsic” reproducibility issues, which we defined as issues originating from hardware and software for deep learning, in deep learning-based MR reconstruction tasks. The sources were tested for training as well as inference of a network. Our study implies that sharing a trained network is recommended to address the intrinsic reproducibility issues.
Methods
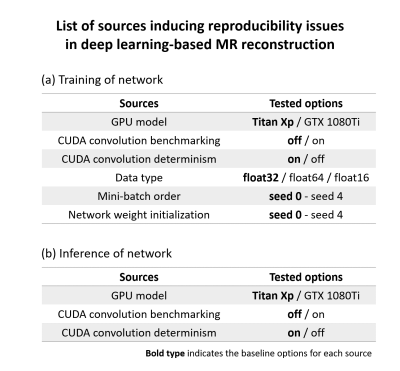
From extensive literature search and investigation, we identified and tested six sources that may cause intrinsic reproducibility issues. They are 1) GPU model, 2) CUDA convolution benchmarking option, 3) CUDA convolution determinism option, 4) data type, 5) mini-batch order, and 6) network weight initialization. The first three sources are related to both training and inference, whereas the last three sources are only related to training because they are not used or varied in inference. This intrinsic reproducibility issue is tested in two kinds of MR reconstruction tasks: quantitative susceptibility mapping (QSM) using QSMnet,2 and parallel imaging using variational network.1 The effect size of each source on the network performance was investigated in QSM whereas key performance difference is confirmed in parallel imaging.[Quantitative susceptibility mapping]
For QSM, the same network architecture and datasets from QSMnet were utilized. To test if the training and inference of the network are reproducible, the network was trained and tested repeatedly using the same computational infrastructure using a single GPU (NVIDIA Titan Xp), the same code implementation using PyTorch,8 CUDA convolution benchmarking on, CUDA convolution determinism off, the same order of mini-batch, and the same network weight initialization values. Then, reproducibility in training was verified by checking training loss for 1000 steps. The reproducibility in inference was verified by comparing the reconstructed test images. After confirming that the reproducibility is guaranteed, the effect size of each source (e.g., GPU model) was investigated by changing the options (Fig. 1; BOLD represents baseline options). For the evaluation of the network performance, NRMSE was measured in the test dataset. A pairwise t-test was conducted for statistical analysis.
[Parallel imaging]
For parallel imaging, dataset1 and codes that are available online were utilized (https://github.com/rixez/pytorch_mri_variationalnetwork). The network was trained five times not using the baseline options in Figure 1. The performances were evaluated using NRMSE. For inference, one network was tested five times, reporting NRMSE in the test dataset. A pairwise t-test was conducted.
Results
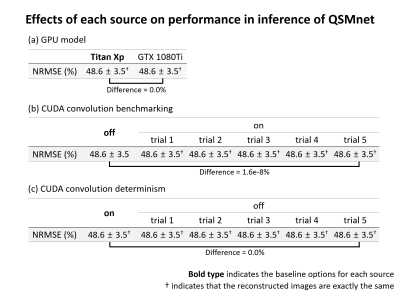
When all six sources were set as the baseline, it was confirmed that the intrinsic reproducibility was guaranteed for both training and inference in QSMnet.Figure 2 shows the effects of each source on performance in the training of QSMnet. For all the sources, the performances of the network were different, suggesting that reproducibility was not guaranteed. In some cases, moreover, the performance differences reached statistically significant differences (e.g., CUDA convolution (Fig. 2b-c), mini-batch order (Fig. 2e), and network weight initialization (Fig. 2f)), demonstrating intrinsic reproducibility is an important issue. For the data type, float64 and float16 were unable to process because of a GPU memory limit and poor precision, respectively. For the reproducibility in the inference of QSMnet (Fig. 3), the results suggest that the GPU model and CUDA convolution determinism have no difference when the options were changed. On the other hand, the CUDA convolution benchmarking had an extremely small performance difference (1.6e-8%).
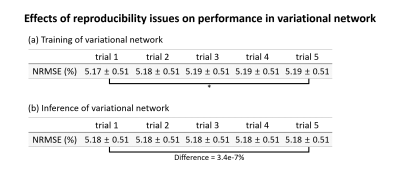
In the variational network (Fig. 4), different training showed differences in performance with a statistical significance whereas the inference had negligible difference (3.4e-7%), confirming our findings from QSMnet.
Discussion and Conclusion
In this work, we demonstrate that the intrinsic reproducibility issues, originating from hardware and software of deep learning, exist in deep learning-based MR reconstruction. Six sources were tested to show the reproducibility issues in training and inference, but additional sources such as OS of the system or deep learning software version may potentially affect the reproducibility. As demonstrated in the QSMnet, the reproducibility issues in training can lead to statistically significant performance differences. On the other hand, the effect size was negligible in the inference of the network. A potential explanation for higher susceptibility to the reproducibility issues in the training process is that it requires more random operations such as mini-batch or network weight initialization. Additionally, floating-point errors may be amplified during training. Therefore, sharing a trained network may address the intrinsic reproducibility issue.Acknowledgements
This work was supported by Creative-Pioneering Researchers Program through Seoul National University(SNU) and by the Korea government (MSIT) (No. NRF-2017M3C7A1047864)References
1. Hammernik, K. et al. Learning a variational network for reconstruction of accelerated MRI data. Magn. Reson. Med. 79, 3055–3071 (2018).
2. Yoon, J. et al. Quantitative susceptibility mapping using deep neural network: QSMnet. Neuroimage 179, 199–206 (2018).
3. Jung, W., Bollmann, S. & Lee, J. Overview of quantitative susceptibility mapping using deep learning: Current status, challenges and opportunities. NMR Biomed. 1–14 (2020) doi:10.1002/nbm.4292.
4. Antun, V., Renna, F., Poon, C., Adcock, B. & Hansen, A. C. On instabilities of deep learning in image reconstruction and the potential costs of AI. Proc. Natl. Acad. Sci. U. S. A. 117, 30088–30095 (2020).
5. Nagarajan, P. et al. The Impact of Nondeterminism on Reproducibility in Deep Reinforcement Learning. 2nd Reprod. Mach. Learn. Work. ICML 2018 9116, 64–73 (2018).
6. Nagarajan, P., Warnell, G. & Stone, P. Deterministic Implementations for Reproducibility in Deep Reinforcement Learning. (2018).
7. Pytorch reproducibility. https://pytorch.org/docs/stable/notes/randomness.html.
8. Paszke, A. et al. PyTorch: An imperative style, high-performance deep learning library. Adv. Neural Inf. Process. Syst. 32, (2019).
Figures