1164
DL2: Robustness of Deep Learning Processing on Deep Learning reconstructed Lumbar Spine MRI
Madeline Hess1, Emma Bahroos1, Misung Han1, Kenneth Gao1, Valentina Pedoia1, and Sharmila Majumdar1
1University of California, San Francisco, San Francisco, CA, United States
1University of California, San Francisco, San Francisco, CA, United States
Synopsis
We demonstrate that fast acquisition images reconstructed robust to segmentation with convolutional neural networks, which are known to generate chaotic errors in response to minimal image perturbations. This finding indicates that low-NEX images with DL-based noise reduction are not only robust to human readers, but machine readers as well.
Introduction
Magnetic Resonance (MR) Imaging is commonly used in clinical care of chronic low back pain, but long scan times are a challenge to scan volume and patient comfort, especially for those with the most pain. Reducing the signal averages, or number of excitations (NEX), can reduce scan time but results in increased signal noise. AIR Recon DL, a deep learning-based algorithm from GE to reduce noise in reduced-NEX (fast) images was previously shown to effectively increase SNR without degrading anatomical structures1. We demonstrate that fast images reconstructed with AIR Recon DL are also robust to segmentation with convolutional neural networks, which are known to generate chaotic errors in response to minimal image perturbations. This finding indicates that low-NEX images with DL-based noise reduction are not only robust to human readers, but machine readers as well.Methods
: A sample of 18 patients (8 female) was retrospectively collected from images acquired clinically with both standard and fast sequences on a GE 3T SIGNA Premier Scanner with an embedded GEM coil array. AIR Recon DL was applied to the k-space data of fast low NEX images at three noise reduction factors, 25% (DL 25), 50% (DL50), 75% (DL75), and 100% (DL100).We leveraged two previously published convolutional neural networks to segment the intervertebral discs and vertebral bodies2 from each T1 sagittal acquisition, both standard and fast at all noise reduction factors. Because images from each acquisition type could not be directly aligned, Sørensen-Dice Similarity could not be used to assess variation in segmentation output. Instead, we assessed the preservation of anatomical features in the images by examining the variation in biomarkers extracted from each segmentation on each sequence: intervertebral disc height and vertebral body volume.
All segmentations were post-processed to smooth, fill holes, and identify largest connected components. Intervertebral disc and vertebral body segmentations were reconstructed from slices into 3D volumes. Intervertebral disc height was calculated by computing a 3D centroid for each disc to find the most central slice, then extracting the shortest side length from a minimum bounding rectangle of the disc segmentation. Vertebral body volume was calculated by computing the sum of all foreground pixels on each body. All metrics were converted to patient-based space using pixel spacing and space between slices.
We treated the standard acquisition as the gold standard, and measured variation in performance of each fast acquisition at each noise reduction factor against this baseline. Variation in performance was measured using mean absolute error and Pearson R correlation.
Results
Sample images from standard and fast acquisitions at each noise reduction factor can be seen in Figure 1. Sample segmentation output from inference on each acquisition type can be seen in Figure 2. Mean absolute errors and Pearson R correlation coefficients are summarized in Tables 1 and 2. Intervertebral disc height on all fast images showed mean absolute errors of 0.794 mm or lower and correlations of 0.798 or higher with low p-values when compared with standard acquisitions. Vertebral body volumes from fast images compared with those of standard images showed mean absolute errors of 4.77 cm3 or lower and correlations of 0.638 or higher with low p-values.Discussion
As seen in Figure 3, metric output from fast images is similar to that of the standard images across noise reduction factors. Many errors were the result of missed segmentations, or correctly segmented regions which were not found in the standard images, as seen in the cluster of points equal zero on either axis. This is likely a feature of the limitations of the segmentation models tested; because the models were not trained to segment all bodies in the field of view, field of view shifts can result in identification of correct anatomy in an undesired location (for example, the uppermost edge of the image see Figure 2). False positives may be a result of the same phenomenon in standard images. Both false negatives and false positives were equally likely to occur across noise reduction factors, suggesting errors emerge because of NEX reduction and are not a feature of the AIR Recon DL algorithm.Conclusion
Fast MR acquisitions of the lumbar spine are a critical step toward providing comfortable and high-quality care for patients with chronic low back pain. Fast quantitative metric extraction to enable large scale studies and data accessibility to the research community is an important component to improving our understanding of chronic low back pain. We demonstrate that automatically extracted intervertebral disc height and vertebral body volume are highly similar between standard and fast acquisitions at various levels of noise reduction. The success of these findings suggests that DL-based noise reduction in fast acquisitions does not cause chaotic error propagation in DL-based segmentation as measured by quantitative metric extraction and can be used to scale to large and multi-institution studies.Acknowledgements
We would like to thank the National Institute of Health and National Institute of Arthritis and Musculoskeletal and Skin Diseases for supporting this work with the NIH/NIAMS UH3AR076724 grant.References
1. Bahroos, E. Deep-Learning Based Image Reconstruction for Lumbar Spine MRI at 3T: Clinical Feasibility. in International Society of Magnetic Resonance in Medicine Annual Meeting 2021 (2021).
2. Hess, M. et al. Deep Learning for Automatic Segmentation and Biomarker Extraction in Lumbar Spine MRI. in Orthopedic Research Society Annual Meeting 2021 (2021).
Figures
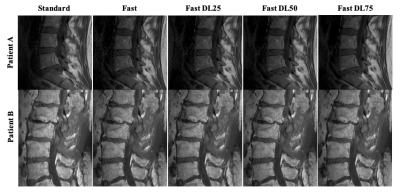
Figure 1: Example images of each acquisition type: standard, fully sampled; accelerated, 100% noise reduction; accelerated, 75% noise reduction; accelerated, 50% noise reduction; accelerated, 25% noise reduction.
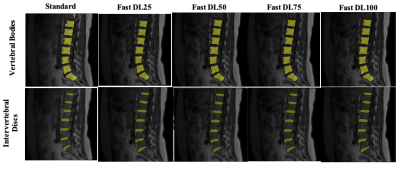
Figure 2: Example segmentations of each model (intervertebral disc, bottom; vertebral body, top) at each noise reduction rate.
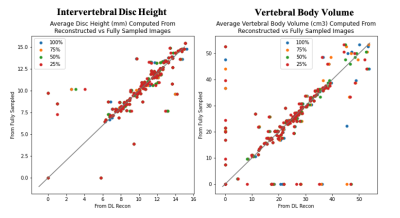
Figure 3: Correlation plot of average biomarker performance in each exam on Deep Learning-reconstructed versus fully sampled images. Each plot is color coded by noise reduction rate.
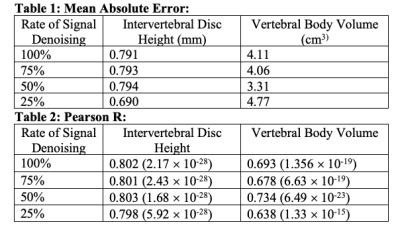
Tables 1 & 2: Mean Absolute Error on each biomarker at each noise reduction factor on each fast image compared with those of the standard images. Pearson R correlation coefficient between metric at each noise reduction factor versus that of the standard images. Correlation coefficient is referenced next to p-value in parentheses.
DOI: https://doi.org/10.58530/2022/1164