1163
Applying advanced denoisers to enhance highly undersampled MRI reconstruction under plug-and-play ADMM framework1Biomedical Engineering, University of Virginia, Charlottesville, VA, United States, 2Radiology & Medical Imaging, University of Virginia, Charlottesville, VA, United States
Synopsis
Accelerating MRI acquisition is always in high demand, since long scan time can increase the potential risk of image degradation caused by patient motion. Generally, MRI reconstruction at higher undersampling rates requires regularization terms, such as wavelet transformation and total variation transformation. This work investigates employing the plug-and-play (PnP) ADMM framework to reconstruct highly undersampled MRI k-space data with three different denoiser algorithms: block matching and 3D filtering (BM3D), weighted nuclear norm minimization (WNNM) and residual learning of deep CNN (DnCNN). The results show that these three PnP-based methods outperform current regularization methods.
Introduction
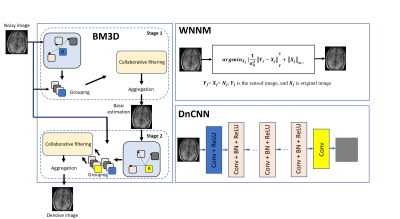
The challenge of fast MRI is to recover the original image from undersampled k-space data. SENSE1 exploits the knowledge of sensitivity maps, and GRAPPA2 uses the learned weighted-coefficients from ACS lines to estimate the missing k-space lines. Compressed sensing3 uses the idea that data can be compressed if undersampled artifacts are incoherent. Therefore, it introduces the concept of sparsity, achieved by regularization terms. L1-ESPIRiT4 also includes regularization terms in soft-SENSE reconstruction to iteratively find the optimal solution. After the PnP prior5 was first proposed by Venkatakrishnan et al, there have been several studies applying this concept to MRI 6,7. Most of these studies focus on the CNN algorithm to complete the denoising process of the PnP algorithm. Alternatively, DnCNN8 may be a better fit. In this work, we explore three advanced denoiser algorithms to reconstruct four-fold undersampled MRI data using the PnP-ADMM framework. The idea behind BM3D9 is that given a local patch, it is not difficult to find many similar patches from nearby. These patches help with denoising, and this idea is typically true for medical images. The human brain, for example, has white matter, gray matter and cerebrospinal fluid, and thus has large nonlocal self-similarity in this sense. WNNM10 aims to improve conventional low rank algorithms by differently weighting singular values in nuclear norm, compared to the general solution which treats singular values equally in order to meet the convex property. Instead of directly outputting a denoised image, DnCNN learns a residual image, and this residual learning and batch normalization could benefit from each other, further improving the denoising performance. This neural network is more natural to combine with the PnP framework.Methods
For MRI reconstruction, the collected signal can be written as: $$s(k_{x},k_{y})=\int_{}^{} \int_{}^{} \rho(x,y)e^{-i2\pi (k_{x}x+k_{y}y)}dxdy,$$which can be written in matrix form as: $$y=Ax+n$$
where $$$y$$$ is the collected data and $$$x$$$ is the expected image; $$$A$$$ represents the sensor matrix; and $$$n$$$ is the noise. In MRI, reconstruction of undersampled data a with regularization term generally is written as follows: $$argmin_{x}||FSx-y||_2^2+\beta(x)$$
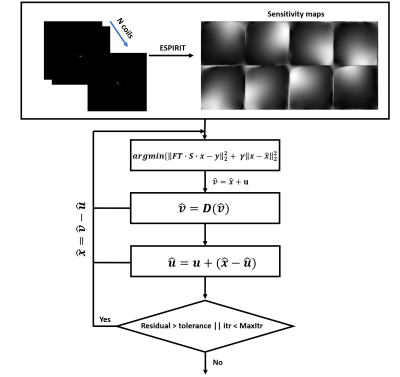
Here, $$$A$$$ is replaced by a sensitivity map weighted operator and a Fourier transform. The PnP-ADMM framework decouples data fidelity and the prior term by splitting variable $$$x$$$ into new variables $$$x$$$, $$$v$$$ and $$$u$$$, and its augmented Lagrangian for MRI reconstruction is $$L_{\gamma}(x,u,v)=||FSTx-y||_2^2+\beta s(v) + \gamma ||x+u-v||_2^2 - \gamma ||u||_2^2$$
Next, we repeat the following steps until convergence according to the ADMM algorithm11, in which $$$\widehat{x}=\widehat{v}-u, \widehat{v}=\widehat{x}+u$$$.
$$\widehat{x}\leftarrow argmin_{x} ||FSx-y||_2^2 + \gamma ||x-\widehat{x}||_2^2 (1)$$
$$\widehat{v}\leftarrow argmin_{v} ||\widehat{v}-v||_2^2 + \gamma s(v) (2)$$
$$\widehat{u}\leftarrow u+(\widehat{x}-\widehat{v}) (3)$$
Here, (1) is a simple MAP estimate of $$$x$$$ with the given data $$$y$$$, and (2) is a denoising process. We could plug in advanced denoising algorithms. In this work, (2) was achieved with BM3D, WNNM and DnCNN.
The flowchart of PnP algorithm is illustrated in Figure 1, and a brief review of the three denoising algorithms is shown in Figure 2. Note that previous methods have used real and imaginary data as the input to the denoising algorithm. Here, we observed that using magnitude data and phase data leads to better performance for the PnP algorithm. BM3D, WNNM and DnCNN algorithms were download online12-14. The tested data was from the NYU fastMRI dataset15, and the displayed brain data and ESPIRIT reconstruction are from Lustig’s ESPIRiT demo16. The undersampling pattern is generated with a variable-density Poisson distribution. Image quality was evaluated with two indexes: peak signal-to-noise ratio (PSNR) and structural similarity index (SSIM), which are defined as follows: $$PSNR=10\cdot log_{10}\frac{max(X)^{2}}{MSE}$$
$$MSE=\frac{1}{mn}\sum_1^n\sum_1^m(X(i,j)-Y(i,j))^{2}$$
$$SSIM=\frac{(2\mu_{x}\mu_{y}+c_{1})(2\sigma_{xy}+c_{2})}{(\mu_x^2+\mu_y^2+c_{1})(\sigma_x^2+\sigma_y^2+c_{2})}$$
where $$$X$$$ is original image and $$$Y$$$ is noisy image with M-by-N matrix size. $$$\mu_{x}$$$and $$$\mu_{y}$$$ are means of images $$$X$$$ and $$$Y$$$ respectively. $$$\sigma_{x}$$$ and $$$\sigma_{y} $$$ are variances of $$$X$$$ and $$$Y$$$, and $$$\sigma_{xy}$$$ represents covariance of $$$X$$$ and $$$Y$$$. $$$c_{1}$$$ and $$$c_{2}$$$ are two variables to stabilize the division with a weak denominator.
Results
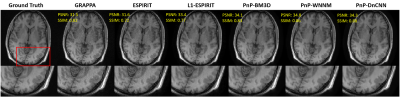
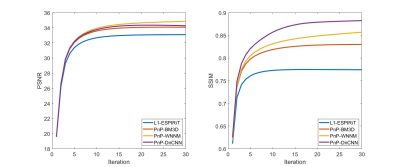
Figures 3 and 4 compare the PSNR and SSIM of the proposed three PnP-based methods and conventional methods with Cartesian data and an acceleration rate R = 4. Even though images reconstructed with GRAPPA suffer from noise, its SSIM is still high compared to ESPIRIT-based methods in which noise was indeed suppressed, but structure similarity decreased. Compared to conventional methods, both PSNR and SSIM of PnP-based methods have been improved: PnP-DnCNN preserved the best structure information while PnP-WNNM achieved highest PSNR.Conclusion
MR reconstruction with advanced denoising algorithms under the PnP-ADMM framework is a flexible approach for image reconstruction. In this initial study, the approach outperformed conventional regularization methods in MRI at acceleration rate R = 4. Image reconstruction with higher acceleration rates in dynamic MRI and a comparison to PnP-CNN are underway.Acknowledgements
We thank Prof. Charles Bouman for his wonderful youtube ECE 641 course. We learnt a lot PnP-related knowledge from his online course.References
[1] Pruessmann KP, et al. MRM 1999; 42:952-962.
[2] Griswold MA, et al. MRM 2002; 47:1202-1210.
[3] Lustig M, et al. MRM 2007; 58: 1182-1195.
[4] Uecker M, et al. MRM 2014; 71:990-1001.
[5] Venkatakrishnan SV, et al. IEEE GlobalSIP 2013; 945-948.
[6] Ahmad R, et al. IEEE SPM 2020; 37:105-116.
[7] Yazdanpanah AP, et al. IEEE ICCV; 2019
[8] Zhang K, et al. IEEE TIP 2017; 26:3142-3155.
[9] Dabov K, et al. IEEE TIP 2007; 16:2080-2095.
[10] Gu SH, et al. IEEE CVPR 2014; 2862-2869.
[11] Boyd S, et al. Found. Trends Mach. Learn. 201; 3
[12] https://webpages.tuni.fi/foi/GCF-BM3D/
[13] https://github.com/csjunxu/WNNM_CVPR2014
[14] https://github.com/cszn/DnCNN
[15] https://fastmri.med.nyu.edu/
[16] https://people.eecs.berkeley.edu/~mlustig/Software.html
Figures