1158
Deep Learning-Based Receiver Coil Sensitivity Map Estimation for SENSE Reconstruction using Transfer Learning1Medical Image Processing Research Group (MIPRG), Dept. of Elect. & Comp. Engineering, COMSATS University, Islamabad, Pakistan
Synopsis
Many Parallel MRI algorithms (e.g. Sensitivity Encoding (SENSE)) require knowledge of the receiver coil sensitivity maps. Magnetic field strength is an important factor in defining the sensitivity maps of the receiver coils in MRI. This paper presents a method to estimate the receiver coil sensitivity maps of a higher magnetic field strength scanner utilizing a deep learning network (denoted as ResU-Net-34), initially trained on the receiver coil sensitivity maps of a lower field strength scanner using transfer learning. SENSE reconstruction results show a successful domain transfer between the receiver coil sensitivities of different magnetic field strengths with the proposed method.
Introduction
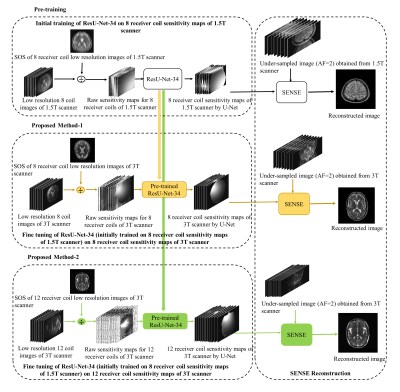
Successful reconstruction of MR images in SENSE1 (and many other pMRI algorithms) is strongly associated with an accurate knowledge of receiver coil sensitivities1. The coil sensitivities change with the magnetic field strength2. A receiver coil array at higher field strength might perform differently than a coil array with an identical geometry at a lower field strength2. Moreover, as the number of receiver coils in an array changes, the sensitivity of an individual receiver coil also changes e.g. an array of 8 receiver coils will have different sensitivity information as compared to an array of 12 receiver coils. This paper presents a new method to estimate the receiver coil sensitivity maps of 3T scanner from a pre-trained ResU-Net-34 via transfer learning. Firstly, the knowledge of sensitivity maps estimation is learnt for an array of 8 receiver coils of 1.5T scanner. Later, the learnt knowledge of sensitivity maps is transferred and fine-tuned for an array of 8 and 12 receiver coils of 3T scanner, separately.Method
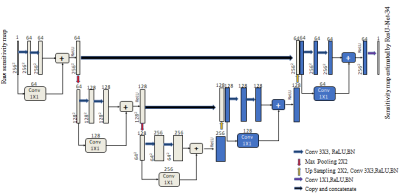
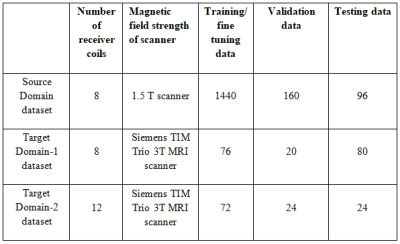
The proposed deep learning framework (Figure 1) is used to estimate the 8 and 12 receiver coil sensitivity maps of 3T scanner via transfer learning. In the proposed methods, the source domain dataset (Table 1) is extracted from human brain Cartesian dataset (of 30 patients of Multiple Sclerosis (MS)) acquired using 1.5T scanner3. The Target Domain-1 dataset (Table 1) is extracted from the human brain Cartesian dataset (of cognitively normal and cognitively declined patients) from Open Access Series of Imaging Studies-3 (OASIS-3)4, which is acquired using 3T scanner. The Target Domain-2 dataset (Table 1) is extracted from human brain Cartesian dataset acquired using 3.0T Siemens Skyra scanner at Case Western Reserve University, Cleveland, OH, USA.Firstly, the customized architecture of ResU-Net-346 (i.e. U-Net integrated with ResNet-34) is trained to generate the sensitivity maps for an array of 8 receiver coils of 1.5T scanner. For training and validation purpose, 1600 simulated images3 from an array of 8 receiver coils of 1.5T scanner are used. The raw sensitivity maps are generated by dividing the low-resolution 8 coil images with the sum of squares image for each dataset1. For label, the sensitivity maps are generated using Eigen value method5 from the corresponding 1600 low-resolution images obtained using 12 auto-calibration signal lines (ACS). Receiver coil sensitivity map is a complex valued data; therefore, the magnitude and phase of the sensitivity maps are trained separately.
Training of the ResU-Net-34 is performed on Python 3.7.1 by Keras using TensorFlow as a backend on Intel(R) core (TM) i7-4790 CPU, clock frequency 3.6GHz, 16GB RAM and GPU NVIDIA GeForce GTX 780 for approximately 7 hours. For training purpose, Adam optimizer with a learning rate of 0.001 is used. The network is trained by using a two-part loss function i.e. Mean Squared Error (MSE) loss and regularization loss (L2-norm). SENSE reconstruction is performed using the estimated coil sensitivity maps by ResU-Net-34 for an array of 8 receiver coils of 1.5T scanner in MATLAB 2018.
In the second phase (i.e. Proposed Method 1), the learnt knowledge of sensitivity maps is transferred and fine-tuned for the sensitivity maps of 8 receiver coils array of 3T scanner4 for 15 epochs. In the third phase (i.e. Proposed Method 2), the learnt knowledge of sensitivity maps is transferred and fine-tuned for the sensitivity maps of 12 receiver coils array of 3T scanner for 20 epochs. In Proposed Methods 1 and 2, end-to-end fine tuning of the pre-trained ResU-Net-34 is performed on Target domain-1 and 2 (Table 1), respectively, using Adam optimizer with a learning rate of 0.0001 (lower as compared to the learning rate used in initial training). SENSE reconstruction (of the under-sampled data (AF=2) acquired from 3T scanner) is performed using ten datasets of 8 channel receiver coil complex sensitivity maps and two datasets of 12 channel receiver coil complex sensitivity maps estimated by Proposed Methods 1 and 2, respectively, in MATLAB 2018.
Results
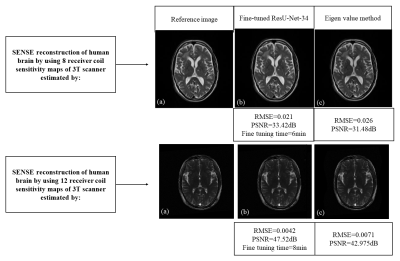
Figure 3 shows SENSE reconstruction results of the uniformly under-sampled (AF=2) human brain datasets by using the sensitivity maps for the 8 and 12 receiver coils array of 3T scanner estimated by the Proposed Methods 1, 2 and Eigen value5 method.Discussion and Conclusion
For any discrepancy between the training and testing datasets, retraining of the neural networks is required from a scratch which may be costly and time consuming. This paper uses the concept of fine tuning for estimating the 8 and 12 receiver coil sensitivity maps of a 3T scanner by utilizing our customized network, initially trained on the 8 receiver coil sensitivity maps of 1.5T scanner. Peak Signal-to-Noise Ratio (PSNR) and Root Mean Square Error (RMSE) values (of the SENSE reconstructed images) show a successful SENSE reconstruction by utilizing the receiver coil sensitivity maps estimated by the proposed methods.Acknowledgements
No acknowledgement found.References
1. Pruessmann, Klaas P., et al. "SENSE: sensitivity encoding for fast MRI." Magnetic resonance in medicine 42.5 (1999): 952-962.
2. Wiesinger, Florian, et al. "Parallel imaging performance as a function of field strength—an experimental investigation using electrodynamic scaling." Magnetic Resonance in Medicine: An Official Journal of the International Society for Magnetic Resonance in Medicine 52.5 (2004): 953-964.
3. Loizou et al. (2011), Multiscale amplitude-modulation frequency-modulation (AM–FM) texture analysis of multiple sclerosis in brain MRI images, IEEE Transactions on Information Technology in Biomedicine 15.1 (2010): 119-129.
4. Pamela J. LaMontagne et al, OASIS-3: longitudinal neuroimaging, clinical, and cognitive dataset for normal aging and alzheimer’s disease, https://doi.org/10.1016/j.jalz.2018.06.1439.
5. Irfan, Amna Shafa, et al. "Sensitivity maps estimation using eigenvalues in sense reconstruction." Applied Magnetic Resonance 47.5 (2016): 487-498.
6. S. Nishank, UNet with ResBlock for Semantic Segmentation, https://medium.com/@nishanksingla/unet-with-resblock-for-semantic-segmentation-dd1766b4ff66.
Figures