1152
Super-resolution mean diffusivity spectroscopic MRI in the human brain1Eunice Kennedy Shriver National Institute of Child Health and Human Development, National Institutes of Health, Bethesda, MD, United States, 2Center for Neuroscience and Regenerative Medicine, The Henry Jackson Foundation, Bethesda, MD, United States, 3National Institute of Neurological Disorders and Stroke, National Institutes of Health, Bethesda, MD, United States, 4Center for Information Technology, National Institutes of Health, Bethesda, MD, United States
Synopsis
We describe a comprehensive pipeline for super-resolution reconstruction of clinical diffusion-weighted MRIs acquired with isotropic diffusion encoding (IDE), or spherical tensor encoding. The pipeline integrates blip-up/down EPI distortion correction and slice-to-volume registration (SVR). Multiple low-resolution IDE-MRIs with different slice orientations relative to the brain are processed to reconstruct high-resolution IDE-MRIs. From high-resolution IDE-MRIs with a wide range of b-values we estimate spectra of subvoxel MD values to describe the distribution of water mobilities in microscopic brain tissue microenvironments. Integrating SVR-reconstruction with IDE is an important step in the clinical translation of MD spectroscopic MRI for fetal MRI applications.
Introduction
The heterogeneous diffusive motions of tissue water in each voxel can be described with an ensemble of microscopic diffusion tensors1-4 with distinct sizes, shapes, and orientations, determined specifically by the local tissue microenvironments sampled by the diffusing water molecules. Mapping spectra of subvoxel mean diffusivity (MD) values, i.e., the size distribution of these microscopic diffusion tensors5, may provide specific clinical information about tissue water mobility in stroke, cancer, brain injury, epilepsy6, and neurodegenerative diseases. We can estimate the spectrum of MDs in subvoxel microenvironments5,7 from multiple DWIs acquired with isotropic8,9 diffusion encoding (IDE), a.k.a. spherical tensor encoding10, over a wide range of b-values (0-4000s/mm2).However, acquiring clinical IDE-MRIs with a sufficiently high signal-to-noise ratio (SNR) and large dynamic range to disentangle multi-exponential signal decays is very challenging. Because averaging complex diffusion MRI signals in vivo is impractical due to shot-to-shot variations in the signal phase induced by physiological motions, the SNR must be increased by using a larger voxel volume, which comes at the cost of lowering the spatial resolution.
Recently, several techniques11-14 have been proposed that use slice-to-volume registration (SVR) to concurrently correct for head motion and reconstruct a 3D volume with high spatial resolution from multiple low-resolution scalar imaging volumes acquired with different relative slice orientations. Since IDE completely removes the effects of both macroscopic and microscopic diffusion anisotropies5, IDE-MRIs can be viewed as scalar imaging volumes, i.e., T2-weighted images, and are therefore well-suited for super-resolution reconstruction using SVR.
We describe a pipeline for SVR-based super-resolution reconstruction of IDE-MRIs with a wide range of b-values needed for MD spectroscopic MRI. By combining SVR with MD spectroscopic MRI we significantly extend the ability to quantify water mobilities in specific tissue microenvironments suitable for important clinical applications such as detecting fetal brain injury in utero.
Methods
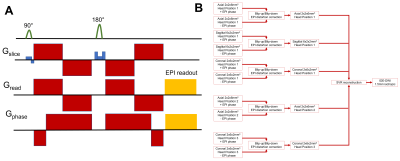
We scanned a healthy volunteer using an MD spectroscopic MRI protocol consisting of IDE scans with 32 different b-values between 0-4000s/mm2. Multiple short (1:45min) whole-brain IDE scans (Fig. 1A) were acquired using a multi-slice EPI acquisition with a cubic FOV of 192x192x192mm3, GRAPPA factor of 2, 96x96 imaging matrix, 32 slices, 2mm in-plane resolution, and 6mm slice thickness. We acquired consecutively 6 IDE-MRI scans with positive (blip-up) and negative (blip-down) EPI phase encoding directions using axial, sagittal, and coronal slice orientations. We repeated these 6 scans for three different head positions relative to the scanner coordinates.The raw IDE-DWIs were reconstructed using the pipeline in Fig. 1B. First, all DWIs in each IDE-MRI scan were corrected for Gibbs ringing15 and B1 variations, denoised16, and rigidly aligned to the first volume in the scan. Next, the multi-b-value IDE scans with opposite EPI phase encoding directions were combined to correct for EPI distortions due to magnetic field inhomogeneities17. Subsequently, the 9 distortion corrected multi-b-value IDE-DWI scans acquired with different slice orientations and head positions were processed using SVRTK11 to reconstruct IDE-DWI volumes at all b-values with a nominal 1.5mm isotropic resolution. Finally, from the super-resolution IDE-DWIs with a wide range of b-values we estimated whole-brain maps of subvoxel MD spectra5,7 and visualized important spectral components.
Results
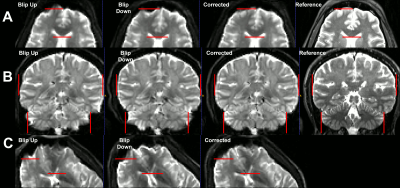
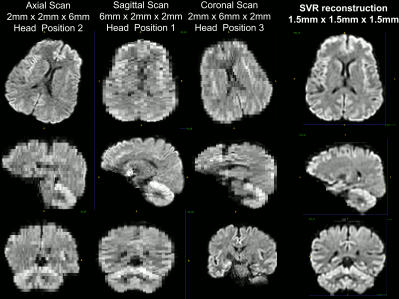
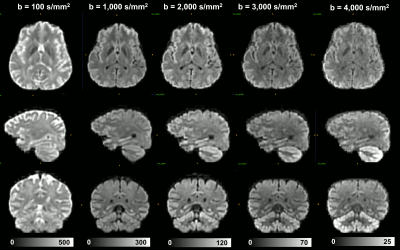
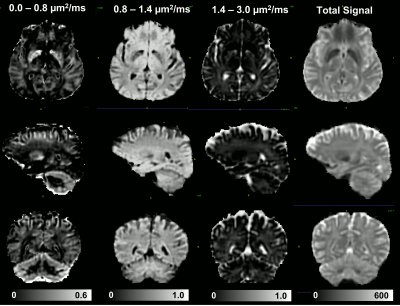
Combining images acquired with positive (blip-up) and negative (blip-down) phase encoding polarity removed orientation-dependent EPI artifacts due to magnetic field inhomogeneities, significantly, providing consistent anatomical accuracy in IDE-DWIs acquired with different slice orientations and head positions (Fig. 2). After SVR reconstruction, the 9 EPI distortion corrected low-resolution multi-b-value IDE-MRI datasets with different slice orientations and head positions were successfully co-registered and combined to obtain corresponding 1.5mm3 DWIs with significantly improved anatomical detail and good SNR even in high-b DWIs (Fig. 3). SVR-reconstructed 1.5mm IDE-DWIs were spatially consistent (co-registered) and showed b-value-dependent brain tissue contrasts in GM, WM, CSF, basal ganglia, and cerebellum (Fig. 4). Subvoxel MD distributions allow clear separation of spectral peaks in the parenchyma and CSF (free water), and reveal a higher fraction of lower MD values in the basal ganglia and cerebellar gray matter, in good agreement with previous studies5,7 (Fig. 5).Discussion
The proposed pipeline corrects for static and dynamic imaging artifacts, i.e., that vary with the relative position of the head such as EPI distortions or B1 inhomogeneities. Combining multiple low-resolution measurements of the same multi-b-value IDE-DWI volumes using various imaging planes and/or head positions yields an effective SNR sufficient for reliable SVR reconstruction of high-resolution multi-b-value IDE data needed for MD spectroscopic MRI. Unlike conventional dMRI, which uses linear tensor encoding2,18, the IDE signal measures diffusion-diffusion correlations very efficiently and allows us to spectrally decompose/disentangle diffusion properties in microscopic tissue domains or environments. Multiband imaging19 could be combined with SVR reconstruction to further accelerate data acquisition for clinical applications in fetal MRI, stroke, cancer, epilepsy, or brain injury.Conclusion
This study represents an important next step in the clinical translation of MD spectroscopic MRI5 and multidimensional MRI7, which promise to provide model-free and tissue-specific assessments of healthy and pathological brain tissues. While other super-resolution techniques are available20,21, the SVR-based methods are particularly well-suited for fetal MRI applications, in which fetal motion can be an intractable problem.Acknowledgements
This work was supported by the Intramural Research Program (IRP) of the Eunice Kennedy Shriver National Institute of Child Health and Human Development, and the CNRM Neuroradiology/Neuropathology Correlation/Integration Core, 309698-4.01-65310, (CNRM-89-9921).References
1 Jian, B., Vemuri, B. C., Özarslan, E., Carney, P. R. & Mareci, T. H. A novel tensor distribution model for the diffusion-weighted MR signal. NeuroImage 37, 164-176 (2007).
2 Topgaard, D. Multidimensional diffusion MRI. Journal of Magnetic Resonance 275, 98-113 (2017).
3 Westin, C. F. et al. Q-space trajectory imaging for multidimensional diffusion MRI of the human brain. Neuroimage 135, 345-362, doi:10.1016/j.neuroimage.2016.02.039 (2016).
4 Magdoom, K. N., Pajevic, S., Dario, G. & Basser, P. J. A new framework for MR diffusion tensor distribution. Scientific Reports 11, 2766, doi:10.1038/s41598-021-81264-x (2021).
5 Avram, A. V., Sarlls, J. E. & Basser, P. J. Measuring non-parametric distributions of intravoxel mean diffusivities using a clinical MRI scanner. Neuroimage 185, 255-262 (2019).
6 Lampinen, B. et al. Tensor-valued diffusion MRI differentiates cortex and white matter in malformations of cortical development associated with epilepsy. Epilepsia 61, 1701-1713, doi:https://doi.org/10.1111/epi.16605 (2020).
7 Avram, A. V., Sarlls, J. E. & Basser, P. J. Whole-Brain Imaging of Subvoxel T1-Diffusion Correlation Spectra in Human Subjects. Frontiers in Neuroscience 15, doi:10.3389/fnins.2021.671465 (2021).
8 Mori, S. & Van Zijl, P. C. M. Diffusion Weighting by the Trace of the Diffusion Tensor within a Single Scan. Magnetic Resonance in Medicine 33, 41-52, doi:10.1002/mrm.1910330107 (1995).
9 Wong, E. C., Cox, R. W. & Song, A. W. Optimized isotropic diffusion weighting. Magnetic Resonance in Medicine 34, 139-143, doi:10.1002/mrm.1910340202 (1995).
10 Lasič, S., Szczepankiewicz, F., Eriksson, S., Nilsson, M. & Topgaard, D. Microanisotropy imaging: quantification of microscopic diffusion anisotropy and orientational order parameter by diffusion MRI with magic-angle spinning of the q-vector. Frontiers in Physics 2, doi:10.3389/fphy.2014.00011 (2014).
11 Kuklisova-Murgasova, M., Quaghebeur, G., Rutherford, M. A., Hajnal, J. V. & Schnabel, J. A. Reconstruction of fetal brain MRI with intensity matching and complete outlier removal. Medical Image Analysis 16, 1550-1564, doi:https://doi.org/10.1016/j.media.2012.07.004 (2012).
12 Gholipour, A., Estroff, J. A. & Warfield, S. K. Robust super-resolution volume reconstruction from slice acquisitions: application to fetal brain MRI. IEEE transactions on medical imaging 29, 1739-1758 (2010).
13 Kim, K. et al. Intersection based motion correction of multislice MRI for 3-D in utero fetal brain image formation. IEEE transactions on medical imaging 29, 146-158 (2009).
14 Shilling, R. Z. et al. A super-resolution framework for 3-D high-resolution and high-contrast imaging using 2-D multislice MRI. IEEE transactions on medical imaging 28, 633-644 (2008).
15 Kellner, E., Dhital, B., Kiselev, V. G. & Reisert, M. Gibbs-ringing artifact removal based on local subvoxel-shifts. Magnetic Resonance in Medicine 76, 1574-1581, doi:10.1002/mrm.26054 (2016).
16 Veraart, J., Fieremans, E. & Novikov, D. S. Diffusion MRI noise mapping using random matrix theory. Magnetic Resonance in Medicine 76, 1582-1593, doi:https://doi.org/10.1002/mrm.26059 (2016).
17 Andersson, J. L. R., Skare, S. & Ashburner, J. How to correct susceptibility distortions in spin-echo echo-planar images: application to diffusion tensor imaging. NeuroImage 20, 870-888, doi:https://doi.org/10.1016/S1053-8119(03)00336-7 (2003).
18 De Swiet, T. M. & Mitra, P. P. Possible systematic errors in single-shot measurements of the trace of the diffusion tensor. Journal of Magnetic Resonance - Series B 111, 15-22, doi:10.1006/jmrb.1996.0055 (1996).
19 Feinberg, D. A. & Setsompop, K. Ultra-fast MRI of the human brain with simultaneous multi-slice imaging. Journal of magnetic resonance 229, 90-100 (2013).
20 Greenspan, H. Super-resolution in medical imaging. The computer journal 52, 43-63 (2009).
21 Souza, A. & Senn, R. in 2008 30th Annual International Conference of the IEEE Engineering in Medicine and Biology Society. 430-434 (IEEE).
Figures