1080
MultiNet CAIPIRINHA: accelerated 1H MRSI with 1-step neural network reconstruction based on augmented MRSI training data1Advanced Imaging Research Center, The University of Texas Southwestern, Dallas, TX, United States, 2The University of Texas Southwestern, Dallas, TX, United States, 3Max Planck Institute for Biological Cybernetics, Tübingen, Germany
Synopsis
We have shown that MultiNet, a neural-network-based image reconstruction, can reconstruct variable-density k-space undersampling schemes to decrease MRSI acquisition times. This used a 4-step method where points are predicted by 4 successively-applied neural-networks off both acquired and previously predicted k-space points. Herein, a 1-step method where points are only predicted off acquired k-space points to reduce reconstruction error was explored. This method was trained using a new augmented MRSI training set and compared to the 4-step reconstruction of new CAIPIRINHA-based schemes and the original schemes. The new 1-step reconstruction method was found to increase SNR and improve metabolic maps.
Purpose
We have shown that MultiNet, a neural-network-based reconstruction, can reconstruct variable-density k-space undersampling schemes to decrease long MR spectroscopic imaging (MRSI) acquisition times (1-3). This used a 4-step method wherein 2-voxel and 1-voxel cross-neighbor and adjacent-neighbor neural-networks (NNs) are used to predict missing points in a step-wise fashion until k-space is filled. Since missing points are predicted off both acquired and previously predicted points, this can result in subpar reconstruction due to compounded error from prior predictions. Consequently, a 1-step method where missing points are only predicted off acquired points was explored herein. To accommodate the variety of distances between missing points and nearest acquired points, the NNs need to be trained on the fully-sampled portion of the MRSI data. For example, two missing points can be 1-point away from the nearest acquired point in two different k-space directions. Thus, the calibration points need one neighboring point in each direction to train both NNs. With other points to predict, the calibration data needs neighboring points in all directions (i.e. fully-sampled). This fully-sampled region is limited by the acceleration rate, however. Thus, a novel augmented training set was created from the MRSI and non-water-suppressed (NWS) data to provide a larger k-space area for optimal NN training. The 1-step method was compared to the original 4-step method with new variable-density CAIPIRINHA sampling schemes and compared to 4-step reconstruction of the originally proposed variable-density sampling schemes (1,2).Methods
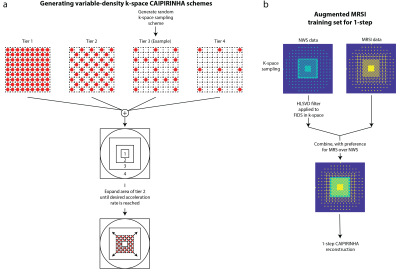
A short-TR/TE elliptically-sampled whole-slice 1H-FID-MRSI sequence was performed in 6 healthy adults (2 male, age:27±2.1 years) on a Philips Achieva 7T scanner with a 32-channel receive head coil with and without water-suppression (non-water-suppressed) (3,4). Parameters include flip angle= 32°, TR/TE=300/1.02ms, 512-points, and a spectral bandwidth=4 kHz from a 10-mm slice at the corpus callosum with a field-of-view=200x200 mm2 and a 64x64 phase-encoding matrix. An anatomical image with 4x4 times the resolution of the MRSI was acquired with the same placement and dimensions using a T1-weighted FFE sequence with flip angle=80° and TR/TE=37/4ms. Data were reconstructed with 20 semi-synthetic anatomical training images and MRSI auto-calibration data (5).Variable-density CAIPIRINHA schemes were generated according to figure 1a. The centermost region was fully-sampled and kept constant while the ratio of the middle-to-outer region was increased until the desired acceleration factor (figure 1a) was reached. This was done to maximize the area that trains the 2-voxel NNs (4-step method) and provide a benign aliasing artifact from the sampling pattern. 1H-MRSI data was retrospectively undersampled while the HLSVD-filtered non-water-suppressed (NWS) data was retrospectively undersampled with complementary k-space masks that had an inverse middle-tier sampling scheme to that of the MRSI and combined to form the augmented MRSI training set (figure 1b). This resulted in a MRSI training set with a large fully-sampled k-space area to train the 1-step NNs (figure 1b).
Lipid removal with L1 regularization was performed (6) and fit with LCModel (24). Signal-to-noise ratio (SNR), lipid RMS, and relative error (RE) in metabolite values between the fully-sampled and accelerated data was calculated as in (1,2) and compared. Student’s t-tests were performed to assess for differences and p-value<0.05 was determined to be statistically significant.
Results
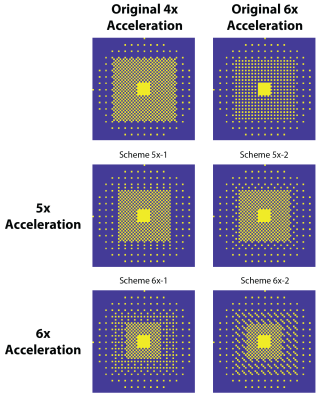
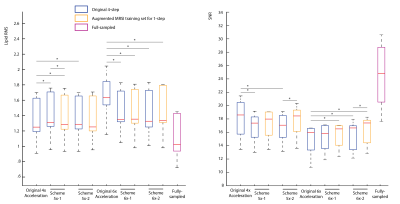
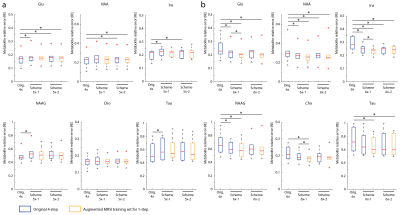
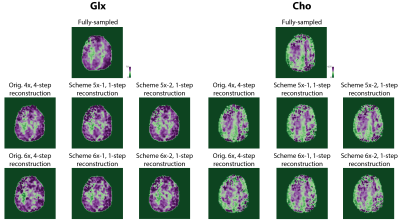
Figure 2 shows the optimized 5-6x sampling schemes and original 4x-undersampling scheme (1) and original 6x-undersampling scheme (2). Lipid RMS and SNR values of the reconstructed spectra are shown in figure 3. Lipid RMS values are 0-4% higher in 5x-1 and 5x-2 relative to the original 4x scheme and all reconstructions of 6x-1 and 6x-2 have equivalent lipid RMS values which are 17-19% lower than the original 6x scheme. SNR is 7-9% higher in the original 4x scheme than in the 4-step reconstructions of 5x-1 and 5x-2 but not the 1-step reconstructions. SNR was 4-8% higher in the 1-step reconstruction of scheme 5x-2 and 6x-2 than the 4-step reconstruction. For 5x-1 and 5x-2, the 1-step method provided the best reconstruction with 3% higher Glu and NAA RE values in 5x-2 and equivalent RE values in 5x-1 relative to the original 4x-undersampling scheme (figure 4). For 6x-1 and 6x-2, the majority of RE values are 13-18% lower than the original 6x-undersampling scheme (figure 4). Glutamate+glutamine (Glx) and Cho maps are shown in Figure 5 where the 1-step reconstructed 5x-1 and 5x-2 maps are similar to the original 4x-undersampling scheme and the 1-step reconstructed 6x-1 and 6x-2 maps are more similar to the fully-sampled data than the original 6x-undersampling scheme.Discussion
The optimized 6x-undersampling schemes significantly reduce lipid artifacts in the brain relative to the original 6x-undersampling scheme and the optimized 5x-undersampling schemes only slightly increase lipid artifacts relative the original 4x-undersampling scheme. Reconstruction with the 1-step method lead to an additional SNR boost due to reduced reconstruction errors. Consequently, metabolic maps can be reconstructed from the 5x-accelerated data without significant information-loss relative to the original 4x-undersampling scheme. Similarly, metabolic maps from the optimized 6x-undersampling schemes contain less error than the original 6x-undersampling scheme. Additionally, the 1-step reconstruction was made possible by the augmented MRSI training set formed from both the NWS and MRSI data, which allowed for a large fully-sampled k-space area for NN training. This allowed the NNs to capture any non-linearities at the edges of k-space.Acknowledgements
This work was funded by the Cancer Prevention and Research Institute of Texas (CPRIT) (Grant Number: RR180056).References
1. Chan et al. ISMRM 2020
2. Nassirpour et al. NeuroImage 2018
3. Chan et al. ISMRM 2021
4. Henning et al. NMR in Biomed 2009
5. Nassirpour et al. NeuroImage 2016
6. Bilgic et al. Magn. Reson. Med 2013
Figures