1073
Reproducibility of multi-slice MR spectroscopic imaging of GABA at 3 T
Helge Jörn Zöllner1, Ipek Özdemir1, Dillip K. Senapati1, Michal Považan2, Georg Oeltzschner1, Kimberly Chan3, Doris D. M. Lin1, and Peter B. Barker1,4
1Russell H. Morgan Department of Radiology and Radiological Science, The Johns Hopkins University School of Medicine, Baltimore, MD, United States, 23 Danish Research Centre for Magnetic Resonance, Hvidovre Hospital, Copenhagen, Denmark, 3Advanced Imaging Research Center, University of Texas Southwestern Medical Center, Dallas, TX, United States, 4F. M. Kirby Research Center for Functional Brain Imaging, Kennedy Krieger Institute, Baltimore, MD, United States
1Russell H. Morgan Department of Radiology and Radiological Science, The Johns Hopkins University School of Medicine, Baltimore, MD, United States, 23 Danish Research Centre for Magnetic Resonance, Hvidovre Hospital, Copenhagen, Denmark, 3Advanced Imaging Research Center, University of Texas Southwestern Medical Center, Dallas, TX, United States, 4F. M. Kirby Research Center for Functional Brain Imaging, Kennedy Krieger Institute, Baltimore, MD, United States
Synopsis
Reproducibility of multi-slice GABA-edited MRSI of the human brain recorded at 3T was investigated in 6 healthy adult volunteers, with and without a retrospective motion compensation scheme applied (MoCo). Overall reproducibility was good with improved performance when MoCo was applied (relative GABA+/tCr difference between visits: 20.8% no Moco, 10.0% with MoCo). Retrospective MoCo is a viable alternative for edited-MRSI when prospective motion correction is unavailable, at least in relatively compliant subject groups.
Introduction
Most edited-MRS studies of GABA in the human brain to date have used single-voxel localization methods (e.g. ‘MEGA-PRESS’)1. MR spectroscopic imaging (MRSI) has the potential to map out the distribution of brain GABA levels, but is technically more challenging to implement. For instance, retrospective, shot-by-shot, phase- and frequency-correction (and outlier rejection) is an important step in the processing of GABA-edited spectra when using single-voxel localization techniques2. When spectral-editing is used in combination with MRSI, retrospective correction (or ‘motion compensation’, MoCo) is more difficult due to the effects of the varying phase-encoding gradients. Recently, a retrospective MoCo method was proposed for single-slice 2D GABA-MRSI at 3T, which compares multiple excitations for each point in k-space3.The purpose of this study was to evaluate the reproducibility of multi-slice 2D GABA-MRSI measurements in the normal human brain at 3T, both with and without retrospective MoCo. It was hypothesized that MoCo would reduce variability and subtraction artifacts.
Methods
6 healthy volunteers (2M/4F, age 28.0±7.7, max 43, min 22) were scanned on 2 occasions between 7-14 days apart (mean ± st.dev = 10±3 days).A 45-minute MR protocol including anatomical MRI, B0 field maps for 2nd-order shim correction, conventional and GABA-edited multi-slice MRSI, and a water reference MRSI was implemented (Philips 3T ‘Ingenia Elition’, 32-channel head coil). The spin-echo edited-MRSI sequence used hypergeometric dualband (HGDB) water and lipid suppression pulses, and 10 outer-volume suppression (OVS) pulses4 (Figure 1A). All MRSI scans were performed with three 15mm oblique-axial slices (2.5 mm gap), 14x17 matrix (elliptical k-space sampling), nominal voxel size 12x12x15 mm (≈2.2 cm3). For GABA-editing, sequence parameters were TR/TE 1.8s/68ms, 4 excitations (2 edit ON, 2 edit OFF), edit ON/OFF frequencies 1.9/0.7 ppm, editing pulse bandwidth 150 Hz (‘sg150’), scan time 22m 21s. Parameters for water MRSI were TR/TE 0.85s/20ms, 1 excitation, scan time 2m 39s. Brain coverage was from the lateral ventricles to the vertex (Figure 1B).
Reconstruction of MRSI data was performed using ‘Osprey’5. MoCo was applied on raw k-space data prior to spatial fast Fourier transformation (FFT) as described previously (3). After spatial transformation, coil combination and phase-correction were applied using information from the water reference scan. Both MoCo and non-MoCo difference spectra were exported into ‘Tarquin’6 for linear combination modeling using the default GABA-edited basis set (including 9 separate Gaussian peaks). The MoCo edit-OFF spectra were also modeled with the ‘1H brain + exp MM’ basis set provided in Tarquin. GABA+/tCr (institutional units, ‘i.u’) was measured for all spectra within the ‘brain mask’ of the central MRSI-slice.
Mean and standard variation of the GABA+/tCr estimates of the center slice were compared between subjects. Additionally, coefficients of variation (CV) were calculated.
Results
Figure 1C shows example difference spectra of the central slice of a representative subject with and without MoCo. MoCo resulted in smaller subtraction artifacts (i.e. residual Cho signal at 3.2 ppm). Figure 2 shows GABA+/tCr maps of the central slice of all 6 subjects at both visits, with and without MoCo. The inter-subject coefficient of variation of mean GABA+/tCr estimates was reduced by from 20 to 16% (visit 1) and from 16 to 11% (visit 2) when MoCo was applied. There was also improved reproducibility between visits - mean relative difference between visits in GABA+/tCr was 20.8% without MoCo, and 10.0% with MoCo (Figure 3). There were no significant differences in GABA+/tCr ratios between visits 1 or 2, with (p = 0.78) or without (p = 0.11) MoCo (paired t-test).Discussion
Multi-slice GABA-MRSI at 3T was found to be reproducible over a 1-to-2-week interval in this study of healthy adult volunteers. Reproducibility was similar to that reported previously for 3D GABA-edited semi-LASER at 3T7. Retrospective MoCo lead to moderate improvements in GABA+/tCr reproducibility; large improvements were not seen, most likely because of the compliant nature of the subjects, and also low B0 drift over the 20min scan time (data not shown). MoCo may be of more value in other groups (e.g. pediatrics) who are more likely to exhibit head motion. While prospective motion correction and associated shim updates and data re-acquisition is likely the best solution for motion sensitive acquisitions such as edited-MRSI8, the required hardware and software is not currently widely available. Retrospective MoCo represents a viable alternative, at least in relatively compliant subjects.Conclusion
3T multi-slice edited-MRSI of GABA in the human brain at a nominal spatial resolution of 2.2 cm3 is feasible, and the use of retrospective motion compensation improves reproducibility.Acknowledgements
Supported by NIH grants R01EB028259 and P41EB031771, DOD W81XWH2010819, R00AG062230, and the Cancer Prevention and Research Institute of Texas (CPRIT, RR180056).References
- Puts NA, Edden RA. In vivo magnetic resonance spectroscopy of GABA: a methodological review. Prog Nucl Magn Reson Spectrosc 2012;60:29-41.2.
- Edden RA, Puts NA, Harris AD, Barker PB, Evans CJ. Gannet: A batch-processing tool for the quantitative analysis of gamma-aminobutyric acid-edited MR spectroscopy spectra. J Magn Reson Imaging 2014;40(6):1445-1452.3.
- Chan KL, Barker PB. Retrospective motion compensation for edited MR spectroscopic imaging. Neuroimage 2019;202:116141.4.
- Zhu H, Edden RA, Ouwerkerk R, Barker PB. High resolution spectroscopic imaging of GABA at 3 Tesla. Magn Reson Med 2011;65(3):603-609.5.
- Oeltzschner G, Zollner HJ, Hui SCN, Mikkelsen M, Saleh MG, Tapper S, Edden RAE. Osprey: Open-source processing, reconstruction & estimation of magnetic resonance spectroscopy data. J Neurosci Methods 2020;343:108827.6.
- Wilson M, Reynolds G, Kauppinen RA, Arvanitis TN, Peet AC. A constrained least-squares approach to the automated quantitation of in vivo (1)H magnetic resonance spectroscopy data. Magn Reson Med 2011;65(1):1-12.7.
- Hnilicova P, Povazan M, Strasser B, Andronesi OC, Gajdosik M, Dydak U, Ukropec J, Dobrota D, Trattnig S, Bogner W. Spatial variability and reproducibility of GABA-edited MEGA-LASER 3D-MRSI in the brain at 3 T. NMR Biomed 2016;29(11):1656-1665.8.
- Bogner W, Gagoski B, Hess AT, Bhat H, Tisdall MD, van der Kouwe AJW, Strasser B, Marjanska M, Trattnig S, Grant E, Rosen B, Andronesi OC. 3D GABA imaging with real-time motion correction, shim update and reacquisition of adiabatic spiral MRSI. Neuroimage 2014;103:290-302.
Figures
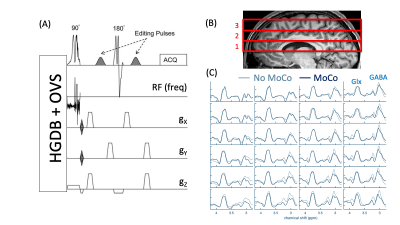
(A) Schematic of the GABA-edited 2D MRSI pulse sequence with presaturation of water and lipid using hyper-geometric dual band (HGDB) pulses and outer-volume suppression (OVS). (B) Sagittal brain MRI showing location of the 3 slices acquired, (C) GABA-edited difference spectra of the central 4x4 grid (middle slice) in one subject with and without MoCo applied. Peaks of GABA and glutamate plus glutamine (Glx) are shown.
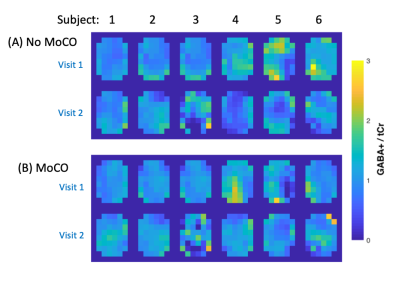
GABA+/tCr MRSI maps from the central slice for all 6 subjects (A) without and (B) with MoCo for visits 1 and 2.
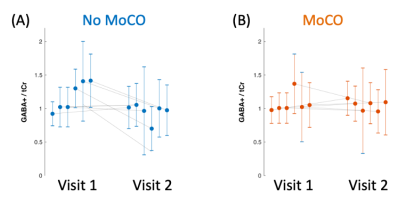
Mean and standard deviation of the GABA+/tCr ratios for all 6 subjects at visits 1 and 2, (A) without and (B) with MoCo.
DOI: https://doi.org/10.58530/2022/1073