1060
Deep learning reconstruction enables accelerated acquisitions with consistent volumetric measurements1GE Healthcare, Calgary, AB, Canada, 2GE Healthcare, Waukesha, WI, United States, 3Cortechs, San Diego, CA, United States, 4GE Healthcare, Menlo Park, CA, United States
Synopsis
This work evaluates brain volumetry measurements obtained from T1w images with fast (PI) and rapid (PI+CS) protocols (< 3 mins) and reconstructed with AIR Recon DL 3D, a deep learning-based reconstruction to reduce noise and ringing. Images were segmented using FDA-cleared software NeuroQuantÒ for quantitative volumetric analysis. Segmentation was successful in all cases and key brain volumes are unchanged between fast and rapid protocols, and further between conventional and DL reconstructions. This work demonstrates that highly accelerated acquisitions and advanced reconstruction methods are suitable for segmentation and volumetric studies and can improve repeatability of measurements.
Introduction
Three-dimensional volumetric T1 weighted sequences, such as magnetization-prepared rapid acquisition with gradient echo (MPRAGE) and variants of Inversion Recovery Spoiled Gradient Recalled Acquisition (IR-SPGR) are core to brain MR exams for lesion assessment. Since they can provide excellent separation of gray matter, white matter, and cerebrospinal fluid, high resolution 3D T1 images are common inputs to tissue segmentation algorithms, such as NeuroQuant [1]. This algorithm provides volumetric measurements and normative ranges of a multitude of brain structures for assessment of various neurological conditions. Previous work demonstrated that accelerated scans with a DICOM-based deep learning (DL) enhancement provided similar results to long unaccelerated protocols [2].Here, we evaluate volumetry accuracy of a reconstruction embedded DL algorithm, AIR Recon DL 3D, which reduces noise and ringing in volumetric images, using both fast and rapid imaging protocols.
Methods
MPRAGE images were acquired using various GE 3T scanners on 10 healthy volunteers (6 F, 4 M; 59.8 ± 11.8 years old) and multichannel receive array coils using fast and rapid protocols. Each protocol was repeated twice, for a total of 4 acquisitions per participant. Both protocols were acquired in the sagittal plane with 1.0 mm3 isotropic resolution, inversion times of 1.0 sec, and bandwidths of 163 Hz/voxel. The fast protocol used a parallel imaging acceleration rate of 2x for a scan time of 5:09; the rapid protocol used a parallel imaging acceleration of 2x and compressed sensing factor of 1.3x for a scan time of 2:56.Images were reconstructed with the conventional vendor reconstruction and with a vendor developed prototype of AIR Recon DL 3D, a volumetric extension of AIR Recon DL 2D [3]. Images were segmented using NeuroQuant. Segmentation quality was assessed visually and volumes of key brain regions, specifically the hippocampus and ventricles were measured.
Results
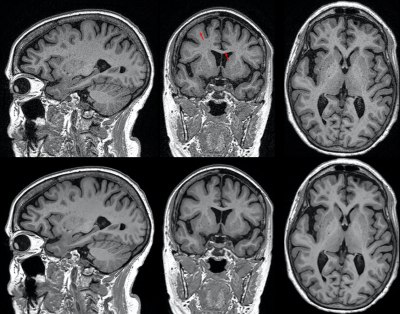
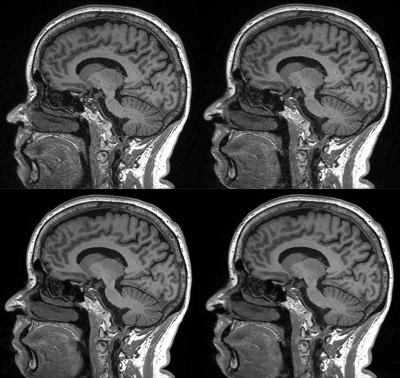
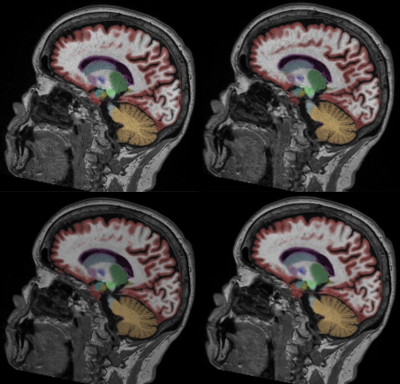
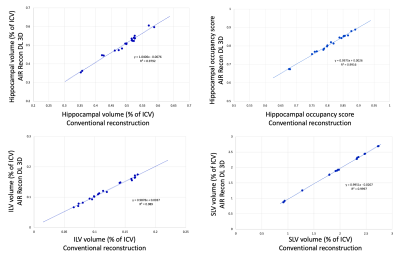
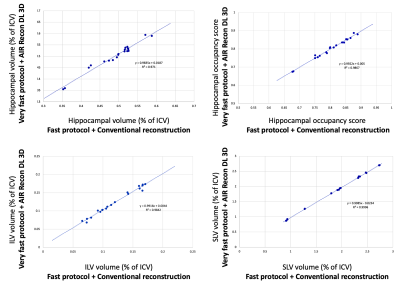
Typical image quality of MPRAGE images with a conventional reconstruction and with AIR Recon DL are shown in Figure 1. Noise is reduced substantially throughout the brain and ringing from truncation artifacts is suppressed. Compared to the conventional reconstruction, subtle structures like the hippocampus and striatum appear more conspicuous with AIR Recon DL 3D. Compressed sensing with AIR Recon DL 3D provides clean and sharp images (Figure 2).Both conventional reconstruction and AIR Recon DL 3D provided visually accurate tissue segmentations (Figure 3). No specific concerns with segmentation were identified in any cases. The use of AIR Recon DL 3D did not significantly alter the volumes measured with NeuroQuant (Figure 4). Both hippocampal volume and occupancy, as well as lateral and superior ventricle volumes were nearly identical with both reconstruction methods. The combination of parallel imaging, compressed sensing, and AIR Recon DL 3D did not compromise volumetry relative to the use of parallel imaging with a conventional reconstruction (Figure 5).
Repeat measurements with AIR Recon DL had a smaller average volumetric differences and standard deviations than did the conventional reconstruction. The average test-retest change in hippocampal occupancy was 0.70% ± 0.54% with a conventional reconstruction and 0.48% ± 0.35% with AIR Recon DL. The inferior lateral ventricle had an average change of 4.2% ± 3.4% with a conventional recon and 3.1% ± 1.6% with DL. The superior lateral ventricle was 0.56% ± 0.45% with conventional recon and 0.37% ± 0.31% with DL.
Discussion and Conclusion
This work demonstrated that AIR Recon DL 3D was able to reduce noise and ringing in images acquired with parallel imaging and with combined parallel imaging and compressed sensing. All images segmented successfully and reported nearly identical volumes of key biomarkers. This work suggests that AIR Recon DL 3D provides the same accuracy for volumetrics as a conventional reconstruction. Furthermore, very short scans are possible while preserving accuracy. The combination of rapid scans and AIR Recon DL 3D shows promise for generating high quality images for both radiological review and for automated volumetry.Acknowledgements
No acknowledgement found.References
[1] Brewer JB, Magda S, Airriess C, Smith ME. Fully-automated quantification of regional brain volumes for improved detection of focal atrophy in Alzheimer disease. AJNR Am J Neuroradiol. 2009 Mar;30(3):578-80. doi: 10.3174/ajnr.A1402. Epub 2008 Dec 26. PMID: 19112065; PMCID: PMC5947999.
[2] Long Wang, Suzie Bash, Sara Dupont, Sebastian Magda, Chris Airriess, Enhao Gong, Greg Zaharchuk, Ajit Shankaranarayanan, and Tao Zhang. Deep Learning Enables Accurate Quantitative Volumetric Brain MRI with 2x Faster Scan Times. 1877, ISMRM 2020.
[3] Lebel, R.M. Performance characterization of a novel deep learning-based MR image reconstruction pipeline. August 2020, http://arxiv.org/abs/2008.06559
Figures