1051
Machine Enhanced Reconstruction Learning and Interpretation Networks (MERLIN)
Kerstin Hammernik1,2 and Thomas Küstner3
1Lab for AI in Medicine, Technical University of Munich, Munich, Germany, 2Department of Computing, Imperial College London, London, United Kingdom, 3Medical Image And Data Analysis (MIDAS.lab), Department of Interventional and Diagnostic Radiology, University Hospital of Tuebingen, Tuebingen, Germany
1Lab for AI in Medicine, Technical University of Munich, Munich, Germany, 2Department of Computing, Imperial College London, London, United Kingdom, 3Medical Image And Data Analysis (MIDAS.lab), Department of Interventional and Diagnostic Radiology, University Hospital of Tuebingen, Tuebingen, Germany
Synopsis
Machine Learning (ML) methods have evolved tremendously during the last decade. A number of frameworks support the development of new ML methods. However, support for high-dimensional and complex-valued data processing is often limited. Therefore, we propose a Machine Enhanced Reconstruction Learning and Interpretation Networks (MERLIN) framework that seamlessly integrates with existing ML solutions such as Tensorflow/Keras and Pytorch and complements them by high-dimensional, complex-valued and MR-specific operators and layers. Furthermore, standard data processing pipelines in Python are provided in the context of MRI reconstruction.
Target Audience
Researchers and developers in the area of machine learning for medical image reconstruction.Purpose
Machine Learning (ML) methods have evolved tremendously during the last decade. In medical imaging, Artificial Intelligence and its subclass ML have the potential to impact all stages of the imaging pipeline, from (1) image acquisition and reconstruction to (2) image analysis and interpretation and (3) diagnosis and prognosis.There exist a number of frameworks, which support the development and production of ML applications1,2, also specifically targeted to MRI3. These frameworks take care of core functionalities, automatic differentiation, have CPU/GPU support, provide the user with a numerous amount of tools to process data, build models, and train/evaluate the models to make the start into ML easier. The two most prominent ones are Tensorflow1 and Pytorch2. While both frameworks now support complex numbers (Tensorflow $$$\geq$$$ v1.0, Pytorch $$$\geq$$$ v1.9), support for complex-valued operations, complex-valued network layers4 and MR-specific operations/layers, e.g. data consistency for MR image reconstruction, are missing.
Furthermore, data loading pipelines are an essential and integral part of the development. Pipelines are usually specific to the underlying data source, layout, application and selected ML framework. Some efforts have been made to unify processing in training and testing such as NiftyNet5 for Tensorflow or TorchIO6 for PyTorch, but each providing unique set of operations that are not directly applicable for high-dimensional complex-valued MR data.
We propose a framework, named Machine Enhanced Reconstruction Learning and Interpretation Networks (MERLIN), that seamlessly integrates with existing Tensorflow/Pytorch implementations and extends these libraries by complex-valued operations. Furthermore, common data pre-processing and data loader functions are provided in Python that complement existing data pipelines, and allows us to use the same data loader transformations independent of the underlying ML framework. MERLIN provides functionalities to process static (2D, 3D) and dynamic (2D+time, 3D+time) data. We also provide a basic tutorial for getting started on ML-based image reconstruction using complex-valued operations.
Functionalities
The proposed MERLIN package is comprised of three modules:- merlinpy (pure Python functions)
- merlintf (Tensorflow/Keras functions)
- merlinth (Pytorch functions)
The MERLIN framework provides (Fig. 1):
- MR-specific data pre-processing: normalization, coil sensitivity map estimation, coil map compression3, $$$\dots$$$
- Data undersampling: variable-density Poisson Disc, Gaussian, VISTA7 $$$\dots$$$
- Data processing pipelines which can be used for Tensorfow and Pytorch and in combination with other data pipelines (NiftyNet, TorchIO, $$$\dots$$$)
- Handling of complex operations
- Handling of multi-dimensional data: 2D, 2Dt, 3D, 3Dt
- Easy switching between complex-valued, 2-channel real-valued or real-valued processing
- Complex layers for convolution, normalization, pooling, activation, $$$\dots$$$
- Data consistency: Proximal mapping, gradient descent, $$$\dots$$$
- Model zoo of commonly used architectures: UNet8, Variational Network9, MoDL10, CINENet11, $$$\dots$$$
Examples
We provide tutorials for image enhancement networks, and physics-based unrolled reconstruction networks for real-valued (magnitude), 2-channel real-valued (real and imaginary component stacked in separate dimension) and complex-valued input data in our online GIT repository. All tutorials can be run directly from Google Colab. We illustrate the usage of MERLIN for real-valued and complex-valued ML-based image enhancement on MNIST in Fig. 2. An example for 5x retrospectively VISTA undersampled 2D cardiac CINE MRI is illustrated in Fig. 3, using MERLIN.The simplicity of running MR-based reconstructions is illustrated in Fig. 4. Conventional forward and backward operators are included in MERLIN which operate for arbitrary input dimensions. Data consistency operators (proximal mapping, gradient descent) can be used within an unrolled reconstruction as data consistency layer or to perform an iterative SENSE reconstruction on the GPU.
Resources
The MERLIN source code is available under: https://github.com/midas-tum/merlinConclusion
We present MERLIN, an ML-based framework for complex-valued, high-dimensional operators and MR-specific data processing and functionalities that integrate with existing backbone frameworks, i.e., Python, Tensorflow/Keras and Pytorch. MERLIN facilitates the workflow to switch between different frameworks, and provides flexibility to integrate further extensions.Acknowledgements
We would like to thank all current and future MERLIN contributors.References
1. Abadi et al. TensorFlow: Large-Scale Machine Learning on Heterogeneous Systems. 2015.2. Paszke et al. PyTorch: An Imperative Style, High-Performance Deep Learning Library. In: Wallach H, Larochelle H, Beygelzimer A, d Alché-Buc F, Fox E, Garnett R, editors. Advances in Neural Information Processing Systems 32: Curran Associates, Inc.; 2019. p 8024-35.
3. Uecker et al. Proc Intl Soc Mag Reson Med 2015.
4. Virtue. Complex-valued deep learning with applications to magnetic resonance image synthesis: University of California, Berkeley; 2019.
5. Gibson et al. Computer Methods and Programs in Biomedicine 2018;158.
6. Pérez-García et al. Computer Methods and Programs in Biomedicine 2021.
7. Ahmad et al. Magn Reson Med 2015;74(5).
8. Hauptmann et al. Magn Reson Med 2019;81.
9. Hammernik et al. Magn Reson Med 2018;79(6).
10. Aggarwal et al. IEEE Trans Med Imaging 2019;38(2).
11. Küstner et al. Sci Rep 2020;10(1).
Figures
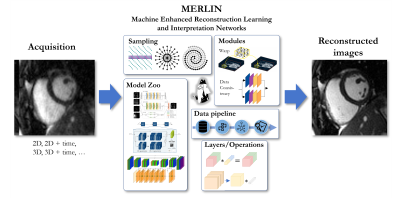
Fig. 1: Machine Enhanced Reconstruction Learning and Interpretation Networks (MERLIN) framework to facilitate development of ML algorithms for MRI.
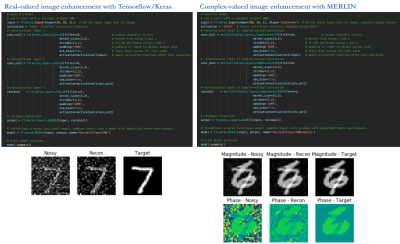
Fig. 2: MNIST code example comparing real-valued (magnitude) image enhancement with layers from Tensorflow/Keras (left) and complex-valued image enhancement with layers from MERLIN. MERLIN is structured the same way as Tensorflow/Keras, allowing for easy switching between the core framework and the extended functionality of MERLIN.
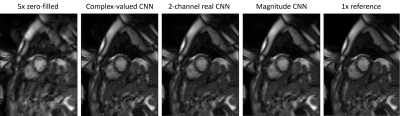
Fig. 3: Exemplary ML-based MR image reconstruction from a 5x retrospectively VISTA7 undersampled 2D cardiac CINE MRI using an image enhancement convolutional neural network (CNN) for real-valued (magnitude), 2-channel real-valued (real and imaginary component stacked in separate dimension) and complex-valued input data.

Fig. 4: Iterative SENSE reconstruction on GPU using forward and backward operators that accept arbitrary input dimensions.
DOI: https://doi.org/10.58530/2022/1051