1043
A modular software “sequence building block” for interleaved MRS and MRI acquisition on 7T Terra1University of Cambridge, Cambridge, United Kingdom
Synopsis
In this work, we present a software “sequence building block” that enables interleaving MRS acquisitions (X-nuclear or 1H) inside a base imaging sequence (typically 1H) on the Siemens 7T Terra platform. We envisage using this approach for dynamic B0 shim and frequency offset correction to mitigate breathing artefacts in 31P-MRSI of heart and liver.
Introduction
Many multinuclear MRI scans use dual tuned coils [1] to facilitate localisation and shimming. We present a software “sequence building block” that enables interleaving MRS acquisitions (X-nuclear or 1H) inside a base imaging sequence (typically 1H) on the Siemens 7T Terra platform. We envisage using this approach for dynamic B0 shim and frequency offset correction to mitigate breathing artefacts in 31P-MRSI of heart and liver.Methods
All data was acquired using a 7T Terra MRI Scanner (Siemens). We tested using three RF coils: a 1Tx/30Rx 1H/13C head coil for 13C phantom acquisitions (Rapid Biomedical, figure 1B), a 2H/1H head coil (Virtumed, figure 1C), and a 31P/1H liver/heart array coil (Tesla Dynamic Coils, built for the EU NICI project and shown in Figure 1D).We presented an early prototype 31P/1H interleaved FID sequence at ISMRM2021 [2]. This year, we have developed this into a robust “Sequence Building Block” (SBB) software module that can be used to interleave MRS into any imaging sequence with only a few lines of code. Our SBB module supports interleaving any pair of nuclei supported by the coil (or interleaving scans on a single nucleus). It supports interleaved FID, SVS-STEAM and CSI-FID acquisitions. Key parameters are available on the sequence special card such as bandwidth, pulse duration, spatial resolution and excitation pulse shape (Figure 1A). The voxel position is set using a “navigator” user interface element in the standard slice selection graphical user interface.
We added our SBB module to the product GRE sequence.
First we acquired 1H images interleaved with 13C-FID with 400 V rectangular pulse with 1s duration, 1s TR for spectroscopy acquisition and 0.5s for imaging using a 1H/13C volume head coil (Rapid Biomedical), on a spherical phantom of 2L containing 100mM of glutamate, 1% of NaN3, and 5g of NaCl (Figure 2A).
Second we acquired 1H dual-echo GRE B0 maps interleaved with 31P-CSI-FID with 500mm FOV, 16x16 matrix size, 40mm slice thickness, 400V sinc pulse, 1s TR for spectroscopy acquisition and 0.5s for imaging using a 1H/31P liver/heart coil (Tesla Dynamic Coils) on a hexagonal prism phantom containing 26L distilled water, 78g NaCl, and 150g H2KPO4 (Figure 2B).
Third we acquired 1H images interleaved with 2H CSI-FID with 250mm FOV, 8x8 matrix size, 40mm slice thickness, 400 V sinc pulse, 1s TR for spectroscopy acquisition and 0.5s for imaging using a 1H/2H head coil (Virtumed) on a cylindrical phantom containing 5.4L water, 162g agar, 4g Dotarem, 5g NaCl, 10g sodium benzoate and 62g deuterium oxide (Figure 2C).
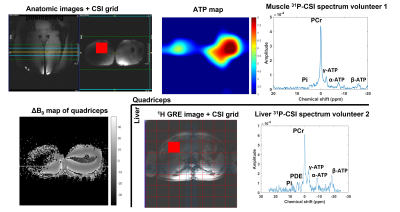
In vivo scans were acquired in quadriceps and liver of healthy volunteers (N=2, male) using the 1H/31P liver/heart coil (Tesla Dynamic Coils). We acquired dual-echo GRE B0 -maps (TE = 3.45ms and 7.39ms) interleaved with a 31P-CSI-FID acquisition (400mm FOV, 8x8 matrix size, 40mm slice thickness, 400 V sinc pulse, 1s TR for spectroscopy acquisition and 0.5s for imaging).
To demonstrate the potential for online B0 frequency correction, phantom scans with interleaved 1H B0-maps and 31P FIDs were acquired while the magnetic field was deliberately disturbed by moving a ferromagnetic object (coin) safely secured inside a plastic bottle near to the phantom.
All data were processed in Matlab (Mathworks, R2019b) using the OXSA toolbox [3] and additional custom functions.
Results
Figure 3 shows the results from the phantoms acquisitions.Figure 4 shows the SNR improvement achieved by correcting the shift in frequency for every transient before averaging.
Figure 5 shows a selection of in-vivo results that illustrate the versatility of our module.
Conclusion
We have developed a software “SBB” module that enables an X-MRS or MRSI interleaved acquisition to be added easily to any imaging sequence. We are happy to share this SBB subject to the usual formalities. To validate our implementation, we ran phantom scans with 1H imaging and interleaved 13C, 31P or 2H MRS. We demonstrated the combined sequence in vivo in quadriceps and liver. Finally, we demonstrated the potential for dynamic 1H-based frequency correction of X-nuclear transients in a phantom scan with deliberate manipulation of the B0 field.Acknowledgements
This project has received funding from the European Union’s Horizon 2020 research and innovation programme under grant agreement No 801075, “NICI”. C.T.R. is funded by the Wellcome Trust and Royal Society (098436/Z/12/B). This research was supported by the NIHR Cambridge Biomedical Research Centre (BRC-1215-20014). The views expressed are those of the author(s) and not necessarily those of the NIHR or the Department of Health and Social Care.References
[1] Choi, Chang-Hoon, et al. "The state-of-the-art and emerging design approaches of double-tuned RF coils for X-nuclei, brain MR imaging and spectroscopy: A review." Magnetic resonance imaging (2020): S0730-725X.
[2] Jabrane Karkouri et. al., Interleaved 31P MRS and 1H dual-echo GRE B0 map pulse sequence for 7T Terra scanners, ISMRM 2021, Abstract ID : 4033
[3] Purvis LAB, Clarke WT, Biasiolli L, Valkovic L, Robson MD, Rodgers CT. OXSA: an open‐source magnetic resonance spectroscopy analysis toolbox in MATLAB. PLoS ONE. 2017;12:e0185356.
[4] Meyerspeer, Martin, et al. Simultaneous and interleaved acquisition of NMR signals from different nuclei with a clinical MRI scanner. Magnetic resonance in medicine 76.5 (2016): 1636-1641.
[5] , , , et al. Interleaved 31P MRS/1H ASL for analysis of metabolic and functional heterogeneity along human lower leg muscles at 7T. Magnetic Resonance in Medicine 2020; 83: 1908– 1919.
Figures
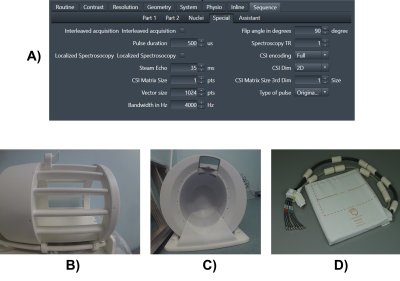
Figure 1: A) The sequence special card enables key parameters to be set for the interleaved SBB module. B) shows the 13C/1H head coil, C) the 2H/1H head coil and D) the 31P/1H liver/heart coil.

Figure 2: The different phantoms for the x-nuclei applications are shown here. (A) 13C phantom, (B) 31P phantom and (C) 2H phantom.
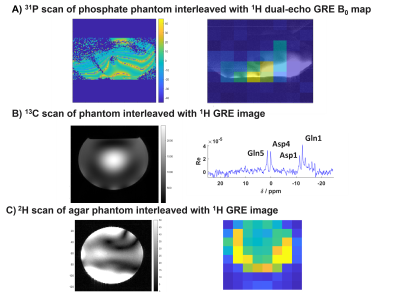
Figure 3: Validation of the SBB interleaved module on different phantoms and different nucleus. In A), we can see the B0-map acquired and interleaved with a 31P-CSI sequence, and the Phosphate map is shown overlayed with the phantom image. In B), the phantom image is shown interleaved with the 13C-FID spectrum and in C), the 2H map resulting from the interleaved 2H-CSI acquisition is shown next to the phantom image.
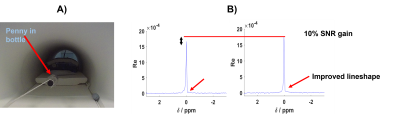
Figure 4: Frequency correction illustration with in A) a picture of the penny in the bottle used to disturb the static magnetic field, and in B) the resulting averaged spectra from the 31P-FID acquisition without frequency correction on top and with frequency correction on the bottom. This can be very helpful for low SNR x-nuclei applications such as 31P MRS. 1H pencil navigators or B0 maps can be acquired and interleaved at the same time for an online and interactive correction which would be beneficial for SNR. Moreover, any 1H transmit pulse will give beneficial NOE boost for 31P signals.