1018
Deep Learning-based Stroke Region Segmentation on Susceptibility Weighted Images in Acute Stroke1Centre for Biomedical Engineering, Indian Institute of Technology, Delhi, New Delhi, India, 2Department of Radiology, Fortis Memorial Research Institute, Gurugram, India, 3Department of Biomedical Engineering, All India Institute of Medical Sciences, New Delhi, India, 4School for Artificial Intelligence, Indian Institute of Technology, Delhi, New Delhi, India
Synopsis
SWI plays a critical role in stroke in demonstration of hemorrhagic transformation of stroke and demonstration of thrombus in the intracranial arteries. Recently it has been used to quantify the penumbra in acute stroke. It highlights venous vasculature in acute stroke due to hypoxia in the acute ischemic tissue without the need for any contrast injection and adding additional sequence that results in time penalty. The objective of this study was to develop an automatic framework for penumbra detection using only SWI images. Evaluation of segmentation results shows a dice similarity coefficient of 0.72 and a jaccard index of 0.60.
Introduction
Acute stroke is a medical emergency in which the blockage of blood flow to the brain tissue leads to tissue damage. Nearly 80% of the strokes are acute ischemic strokes1. CT and MRI are the most common imaging modalities used to detect a stroke. Among MRI - Diffusion-Weighted-Imaging (DWI) and Perfusion-Weighted-Imaging (PWI) sequences are used to identify the stroke core and the penumbra region2–4. Recent studies have shown that Susceptibility-Weighted-Imaging (SWI) can be used to determine the stroke area along with DWI and PWI5–8. The precise extraction of extent of salvageable tissue is essential to prevent the extent of tissue damage and its impact on normal brain function. Manual segmentation of the lesion is time-consuming and also subjective to both inter-and intra-observer variability. In recent times, convolutional-neural-networks (CNN) based deep learning models have shown high segmentation accuracy, outperforming standard segmentation approaches. Due to their ability to extract complex features, deep learning models may segment stroke core and penumbra from DWI and PWI images9–12. This study aims to develop automatic segmentation tool to segment penumbra from SWI images which are routinely obtained in stroke patients with out wasting additional time on data acquisition to obtain perfusion imaging.Materials and Methods
MRI Data Acquisition:MRI data of 24 patients (age: 56.87 ± 19.06 years) with large territory acute ischemic stroke was used in this study. Imaging was performed on a 3.0T MRI scanner (Ingenia, Philips Healthcare, The Netherlands). MRI acquisition sequences included 2D FLAIR, DWI, 3D SWI, 3D pCASL, and 3D Time-of-Flight MRA.
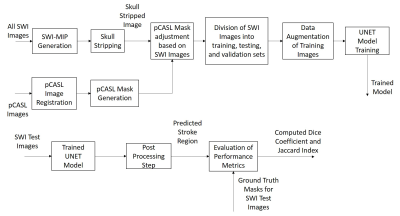
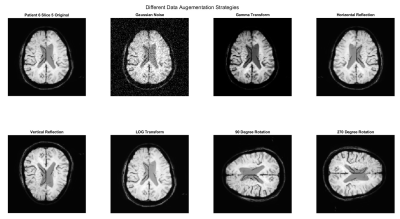
Data Processing: SWI magnitude images and manually annotated pCASL masks were used for developing the proposed methodology. pCASL images were used to generate masks since they provide higher contrast in the stroke region. Manual annotations on pCASL images were performed by an experienced radiologist. Since SWI magnitude images were acquired at 1mm slice thickness (no interslice gap) and pCASL images were acquired at 6mm slice thickness (no interslice interval), SWI minimum-intensity (SWI-MIP) maps were generated using the algorithm proposed previously8. All the acquired images were registered with SWI-MIP images using affine co-registration. The images were resized to 128by128 pixels. Skull stripping on SWI images for alignment of pCASL mask with the brain region was performed using an in-house developed algorithm. Only slices containing the stroke region were considered in this methodology. The complete flowchart of the methodology is shown in figure 1. A total of 148 slices from 24 patients were obtained. These slices were randomly distributed into training, validation, and testing set in the ratios of 0.64, 0.16, and 0.20. Data augmentation strategies like the addition of Gaussian Noise, Gamma Transform, Log Transform, Image Rotation, and Reflection were performed on the training set to prevent overfitting. UNET is one of the most common models for image segmentation and can provide good results even with a small number of datasets13. The UNET was trained for 100 epochs on the training set. The following parameters were optimized; optimizer, batch size, activation functions, initializer, data augmentation strategies, the effect of batch normalization, loss function, and L2 Regularization. A post-processing step was performed to eliminate the false positives.
Results
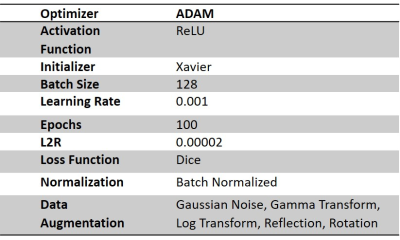
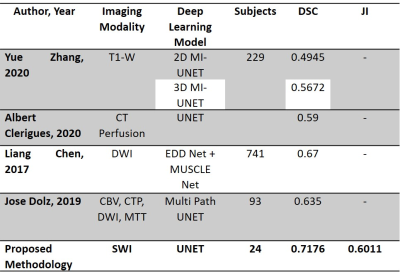
Training the UNET model on SWI images and pCASL ground truth resulted in a dice coefficient (DSC) of 0.72±0.23 and Jaccard index (JI) of 0.60±0.24. During the optimization process, it was identified that the model performed best using ADAM optimizer with a batch size of 128. All data augmentation strategies, as shown in figure 2, were implemented as mentioned above. It was also determined that batch normalization was an essential step as both the DSC and JI values were adversely affected in the absence of batch normalization. Table 1 shows the optimized parameters of the UNET. Table 2 shows the segmented regions and their comparison with the ground truth pCASL image. Table 3 shows that the proposed methodology provides reasonably better stroke lesion segmentation than existing state-of-art methods.Discussion
The study evaluated the stroke lesion detection performance of SWI images using UNET. The existing state-of-art models use conventional MRI imaging sequences like T1W, DWI and PWI. Also, some of the models have not implemented data augmentation strategies. In some models, the batch size is set to 8. Studies have reported that a small batch size leads to more significant errors in segmentation than larger batch size. In order to reduce this error, the batch size has been set to 128. From table 3, it is evident that the proposed methodology has provided reasonably good accuracy (DSC: 0.72, JI: 0.60) than the existing deep learning models trained on DWI and PWI images. This study highlights that SWI images can be used to identify stroke lesions with reasonable accuracy. However, these are preliminary results, and the model needs to be validated on large sample sizes.Conclusion
The study suggests that a deep learning model trained on SWI images can identify the stroke penumbra alone with improved accuracy. The trained model achieved a DSC value of 0.72 and a JI value of 0.60, better than existing state-of-art stroke lesion segmentation models.Acknowledgements
This work is supported by IIT Delhi, India, and Fortis Memorial Research Institute, Gurugram, India.References
1. Norrving B: Stroke And Serebovascular Disorders. Oxford Univ Press 2014; 53:9.
2. Xu K, Gu B, Zuo T, et al.: Predictive value of Alberta stroke program early CT score for perfusion weighted imaging - diffusion weighted imaging mismatch in stroke with middle cerebral artery occlusion. Medicine (Baltimore) 2020; 99:e23490.
3. Lassalle L, Turc G, Tisserand M, et al.: ASPECTS (Alberta Stroke Program Early CT Score) Assessment of the Perfusion-Diffusion Mismatch. Stroke 2016; 47:2553–2558.
4. Chen F, Ni Y: World Journal of Radiology. 2012; 4:63–74.
5. Wu X, Luo S, Wang Y, et al.: Use of susceptibility-weighted imaging in assessing ischemic penumbra. :4–7.
6. Wang T, Zhu L, Hu C, et al.: The diagnostic value of susceptibilityweighted imaging for ischemic penumbra in patients with acute ischemic stroke. Technol Heal Care 2017; 25:S449–S457.
7. Luo S, Yang L, Wang L: Comparison of susceptibility-weighted and perfusion-weighted magnetic resonance imaging in the detection of penumbra in acute ischemic stroke. J Neuroradiol 2015; 42:255–260.
8. Bhattacharjee R, Gupta RK, Das B, Dixit VK, Gupta P, Singh A: Penumbra quantification from MR SWI-DWI mismatch and its comparison with MR ASL PWI-DWI mismatch in patients with acute ischemic stroke. NMR Biomed 2021; 34:1–15.
9. Zhang Y, Wu J, Liu Y, Chen Y, Wu EX, Tang X: MI-UNet: Multi-Inputs UNet Incorporating Brain Parcellation for Stroke Lesion Segmentation from T1-Weighted Magnetic Resonance Images. IEEE J Biomed Heal Informatics 2021; 25:526–535.
10. Clèrigues A, Valverde S, Bernal J, Freixenet J, Oliver A, Lladó X: Acute and sub-acute stroke lesion segmentation from multimodal MRI. Comput Methods Programs Biomed 2020; 194.
11. Chen L, Bentley P, Rueckert D: Fully automatic acute ischemic lesion segmentation in DWI using convolutional neural networks. NeuroImage Clin 2017; 15(May):633–643.
12. Dolz J, Ben Ayed I, Desrosiers C: Dense multi-path u-net for ischemic stroke lesion segmentation in multiple image modalities. Lect Notes Comput Sci (including Subser Lect Notes Artif Intell Lect Notes Bioinformatics) 2019; 11383 LNCS:271–282.
13. Weng W, Zhu X: INet: Convolutional Networks for Biomedical Image Segmentation. IEEE Access 2021; 9:16591–16603.
Figures