0987
VERDICT MRI for estimation of intracellular volume fraction and cell radius: comparison with histology in a mouse model of neuroendocrine tumor1Medical Radiation Sciences, University of Gothenburg, Gothenburg, Sweden, 2Medical Physics and Biomedical Engineering, Sahlgrenska University Hospital, Gothenburg, Sweden
Synopsis
VERDICT MRI provides estimates of intracellular volume fraction and cell radius non-invasively which may facilitate e.g., tumor grade classification and longitudinal studies without the need for biopsy. Tumors of a human SI-NET animal model were irradiated and measured with diffusion MRI. Colormaps of cell radius index and intracellular fraction were derived from both VERDICT analysis of the MR data and histological analysis of stained tumor slices. VERDICT maps of intracellular fraction corresponded well with histology in necrotic tissue, however the cell radius index was poorly estimated in these regions. Further work is needed to optimize VERDICT for different tissue types.
Introduction
Measuring the microstructure of cancer tissue can provide valuable information of the tumor such as malignancy grade and the effect of treatment on the tissue. The common method for tumor tissue analysis today is microscopy examination of samples extracted via biopsy. However, this method only provides information of the limited region of the extraction site and may thus lead to misclassification of the tumor. Furthermore, it poses substantial limitations to the accuracy of longitudinal studies.Vascular, Extracellular and Restricted Diffusion for Cytometry in Tumors (VERDICT) is a mathematical model designed to be fitted to diffusion weighted data acquired at different diffusion times to estimate microstructural parameters such as cell radius and intracellular volume fraction in tumor.1 VERDICT therefore holds potential in providing non-invasive biomarkers for tumor grading and treatment response.
The aim of this study was to compare VERDICT estimations of the cell radius and intracellular volume fraction with histological analysis in a mouse model of human neuroendocrine tumor.
Subjects and methods
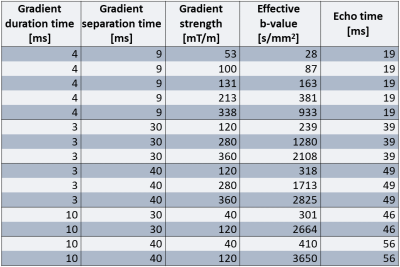
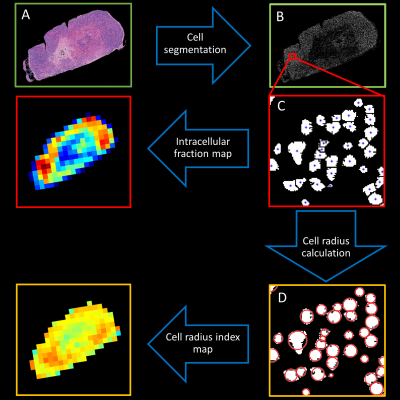
Analysis of VERDICT parameters was done using female BALB/c nude mice (n = 5) of the human SI-NET model GOT1. To provide a more heterogenous tissue the tumors were irradiated externally to an absorbed dose of 8 Gy. 15 days after treatment diffusion weighted MR images of the tumors were acquired using a protocol designed for VERDICT analysis (Table 1) on a 7T system (Bruker,Biospec,MRI GmbH,Ettlingen,Germany). The pixelsize was 400×400μm2 and the slice thickness was 500μm.The VERDICT model was fitted to diffusion MRI data using the AMICO framework2 to estimate the cell radius index, RVERDICT, and intracellular volume fraction, fVERDICT. To make the fitting more robust the diffusion coefficient of the intracellular, extracellular extravascular, and vascular space, as well as the velocity dispersion of the blood flow were fixed as 1×10-9m2/s, 1.5×10-9m2/s, 1.75×10-9m2/s, and 0.6x10-3 m/s respectively. Animals were euthanized immediately following the MRI scans and tumors were extracted and fixated. Slices of the tumors were extracted from the same plane as the MRI images and stained with hematoxylin/eosin to colorize cell nuclei and cell plasma. High resolution images (0.25×0.25μm in-plane) of the stained slices were acquired using light microscopy and cell nuclei were segmented using the software HALO (IndicaLabs,New Mexico,USA). Maps of cell radius index, RHIST, and intracellular area fraction, fHIST, were generated from the stained slices as outlined in Figure 1. RHIST was calculated for each cell as
$$ R_{HIST}=\sqrt{A/π} $$
where A was the area of the cell. The histology maps were downscaled to a resolution similar to that of the MRI images to facilitate comparison.
This study was approved by the Gothenburg Ethical Committee on Animal Research.
Results and discussion
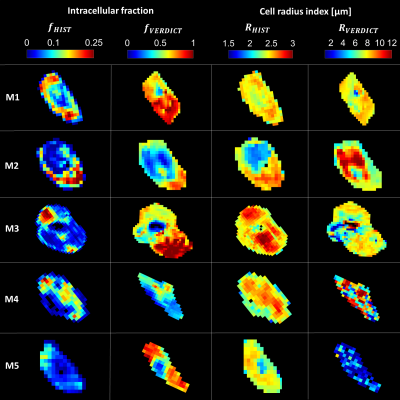
The contrast of the fHIST and fVERDICT maps showed a moderate spatial match overall (Figure 2). Necrotic tissue, seen as regions of low fHIST in the histology maps, were especially well indicated by VERDICT as regions with low fVERDICT. However, maps of RHIST and RVERDICT did not match as well, especially for mouse 4 and 5 where substantial heterogeneity was seen in the VERDICT maps which was not evident in the histology maps.RHIST was consistently lower in necrotic regions, as opposed to RVERDICT which showed higher values (e.g. mouse 2, center region of the tumor). The low intracellular SNR of these regions likely made the estimation of RVERDICT difficult for the VERDICT model. The regularization used in the fitting method may therefore have caused a systematic overestimation of RVERDICT in these areas. This shows one of the limitations of the VERDICT model and can lead to misinterpretation of the underlying microstructure.
The absolute values of intracellular fraction and cell radius index did not match well between histology and VERDICT. Compared to histology, the cell radius index was estimated as 2 – 3 times higher and the intracellular fraction as 4 – 5 times higher when estimated by VERDICT. In the histological evaluation we found that the cell segmentation was not optimal in some areas which lead to underestimation of fHIST. Furthermore, the tumor slices used in the histology analysis were 3 – 4 µm thick and therefore only covered a cross section of the cells. This was not accounted for and, as such, caused underestimation of both RHIST and fHIST.
Mouse 3 showed a region of high fVERDICT in the lower part of the tumor which was not apparent in the histology map. On closer inspection of the histology images this region showed large amount of scar tissue which may have confounded the estimation of fVERDICT.
Conclusion
Estimates of fVERDICT show promise in differentiating necrotic regions from viable regions in the studied mouse model. RVERDICT is likely to suffer from poor accuracy when fVERDICT is low and should be omitted from maps of such regions. Additionally, some tissue types, such as scar tissue, may confound the estimation of microstructural parameters by VERDICT and lead to misinterpretation of the microstructure. Further work is needed to optimize VERDICT for different tissue types and to improve the method used for biological validation of its estimated parameters.Acknowledgements
We are grateful to Emman Shubbar for skillful assistance with animal handling and treatment procedures.
This study was funded by grants from the Swedish Cancer Society, the Swedish Research Council, the King Gustav V Jubilee Clinic Cancer Research Foundation, BioCARE – a National Strategic Research Program at the University of Gothenburg, the Swedish state under the agreement between the Swedish government and the county councils, the ALF agreement, the Sahlgrenska University Hospital Research Funds, the Assar Gabrielsson Cancer Research Foundation, the Adlerbertska Research Foundation, the Herbert & Karin Jacobsson Foundation, the Royal Society of Arts and Sciences in Gothenburg (KVVS), and the Wilhelm and Martina Lundgren Research Foundation.
References
1. Panagiotaki E, Walker-Samuel S, Siow B, et al. Noninvasive quantification of solid tumor microstructure using VERDICT MRI. Cancer Research. 2014;74(7):1902-1912.
2. Bonet-Carne E, Johnston E, Daducci A, et al. VERDICT-AMICO: Ultrafast fitting algorithm for non-invasive prostate microstructure characterization. NMR in Biomedicine. Published online October 31, 2018:e4019.
Figures