0967
Creating a multi-centre harmonised surface-based MRI dataset for the Multi-centre Epilepsy Lesion Detection Project1Great Ormond Street Institute for Child Health, UCL, London, United Kingdom, 2Institute of Computational Biology, Helmholtz Zentrum München, Munich, Germany, 3Penn Statistics in Imaging and Visualization Center, University of Pennsylvania, Philadelphia, PA, United States, 4UCL, London, United Kingdom, 5Wellcome Centre for Human Neuroimaging, UCL, London, United Kingdom
Synopsis
The Multi-centre Epilepsy Lesion Detection (MELD) project presents a methodology to harmonise a large heterogenous cohort of surface-based MRI data. Structural features were extracted from T1w and FLAIR images and pre-processed to reduce systematic site, scanner, and age-specific biases. The harmonised dataset enabled the characterisation of subtle radiological markers of focal cortical dysplasia (FCD), a cortical abnormality causing drug-resistant epilepsy. Machine-learning algorithms trained on the harmonised dataset improved the classification of FCD histopathologies. With open-source protocols and code, the MELD preprocessing pipeline offers a reproducible method to prepare large heterogeneous datasets for statistical analysis and machine-learning tasks.
Purpose
Robust statistical analyses and training for machine-learning tasks in medical imaging requires large datasets, which can be a challenge in rarer pathologies. One solution is data-sharing initiatives involving multi-centre collaborations. However, systematic scanner differences can introduce site-specific biases in the data. Furthermore, heterogeneous patient cohorts mean that imaging features can be affected by developmental effects such as age, as well as the biological variable of interest. To overcome these limitations, careful pre-processing of the data is needed. In this abstract, we present a pipeline to harmonise multi-centre surface-based MRI data in focal cortical dysplasia (FCD), a cause of drug-resistant epilepsy. We then demonstrate how harmonisation of data enables analysis of FCD histopathological subtypes and effective training of machine-learning algorithms to differentiate them.Methods
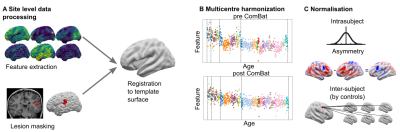
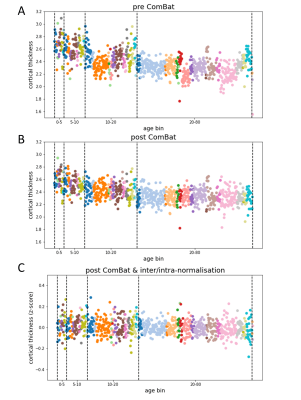
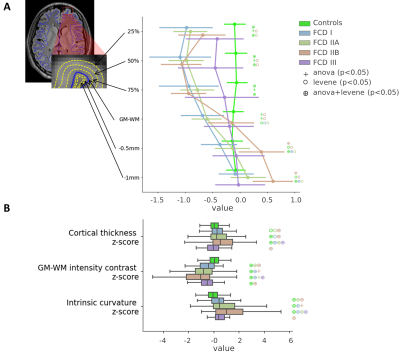
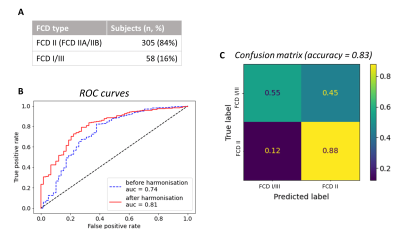
The Multi-centre Epilepsy Lesion Detection (MELD) project collated a cohort of 558 patients with drug-resistant FCD (38 with FCD type I, 114 FCD IIA, 191 FCD IIB, 20 FCD III and 195 without histopathology available) and 379 controls from 20 epilepsy centres worldwide. Participants were included if they were over 3 years old, had been scanned with a 1.5T or 3T MRI and had a T1-weighted scan available. 242 subjects also had a FLAIR scan. Demographic information, such as age and gender, were retrieved. For patients, a manual lesion mask was available. FreeSurfer was used to create reconstructions of the cortical surfaces. Surface-based morphological features (e.g. cortical thickness) and intensity features (e.g FLAIR intensity) were extracted from T1-weighted and FLAIR images by each collaborating site following open-access protocols available on Protocols.io [https://www.protocols.io/researchers/meld-project/protocols]. Features were smoothed, and harmonised across sites and scanners strength using ComBat1, while controlling for age, sex and disease status. Features were then normalised to reduce inter-subject and interregional morphological differences. Inter-hemispheric asymmetry maps of features were created subtracting right hemisphere vertex values from left hemisphere values and vice versa. Normalised cortical thickness and FLAIR intensity distribution at different cortical depths were plotted in four different FCD histopathologies in lesional vertices and paired regions from controls. ANOVA and Levene statistical tests tested for differences among groups’ means and variances, with a significance level set at 5% level. A multi-layer perceptron (MLP) algorithm was trained using morphological and intensity-based MRI features alongside lesion characteristics, e.g. lesion size and location, to differentiate FCD I and III (FCD I/III) from FCD IIA and IIB (FCD II) using 10-fold cross-validation. Receiver operator characteristic (ROC) curves and confusion matrices were computed to evaluate the classifier performances when trained on features before the preprocessing pipeline and after.Results
11 morphological and intensity surface-based features were extracted from T1-weighted and FLAIR images, by each site and pre-processed (Figure 1). Prior to Combat harmonisation, inter-site differences confounded biological variability. For example, in Figure 2A, the biological decrease in cortical thickness with age is partially obscured by inter-site differences in cortical thickness. However, after Combat harmonisation, site-variability is reduced and only biological variability with age remains (Figure 2B). Intra- and inter-subject normalisation adjust for the effects of age and regional variability (Fig 2C). Post-harmonisation, imaging features significantly differ between patients and controls. Furthermore, cortical thickness, intensity contrast at the grey matter / white matter boundary (GM-WM) and intrinsic curvature significantly differ between FCD type I/III and FCD type IIA/IIB (Figure 3). There is a significant reduction in FLAIR intensity for FCD type IIB in the white matter (-1mm cortical depths) not seen in other FCD-subtypes. The MLP algorithm to classify FCD histopathologies had improved performance when trained on pre-processed features than on raw features, with areas under the curve (AUC) of 0.81 and 0.74 respectively (Figure 4B). At optimal performance, the MLP accurately classified 88% of FCD II and 55% of FCD I/III (Figure 4C).Discussion
The MELD project presents a pipeline to harmonise heterogeneous multi-centre data. Features were pre-processed to reduce site, scanner, age, and cortical region-based biases in the data. This pre-processing enables the quantification of subtle MRI feature differences between FCD histopathological subtypes, such as the FLAIR hypo-intensity in the white matter for FCD type IIB, reflecting the transmantle sign only visible in this histopathology. Moreover, these features can then be used as inputs for machine-learning algorithms to differentiate FCD histopathologies. This processing pipeline is open source and available at github.com/MELDproject.Conclusion
We present an open-source framework to harmonise large multi-centre MRI datasets. We demonstrated its utility to characterise subtle differences in MRI features between histopathological subtypes in FCD. This framework is an example of how well-powered quantitative analyses and machine learning tasks can be carried out on medical imaging of rarer pathologies.Acknowledgements
The MELD project is funded by the Rosetrees Trust. A full list of collaborators can be found at https://meldproject.github.io//docs/collaborator_list.pdfReferences
1. Fortin JP, Nicholas Cullen, Yvette I. Sheline et al. 2018. “Harmonization of Cortical Thickness Measurements across Scanners and Sites.” NeuroImage 167 (February): 104–20.Figures