0966
Predicting changes in B0 field due to patient movement dynamically for MRI via Deep Learning1High Field MR Center, Department of Biomedical Imaging and Image-guided Therapy, Medical University of Vienna, Vienna, Austria, 2Computational Imaging Research Lab, Department of Biomedical Imaging and Image-guided Therapy, Medical University of Vienna, Vienna, Austria, 3Department of Neurosurgery, Medical University of Vienna, Vienna, Austria, 4High Field MR Center, Center for Medical Physics and Biomedical Engineering, Medical University of Vienna, Vienna, Austria, 5Christian Doppler Laboratory for Clinical Molecular MR Imaging, Medical University of Vienna, Vienna, Austria
Synopsis
We proposed a U-net-based neural network to predict changes in B0 field due to patient movement. The network was trained and tested on 7T in vivo volunteer data. The method was compared against two other approaches. The estimated B0 maps were compared against the ground truth. The results suggest that prediction of B0 maps is feasible and by combination with external tracking a considerable improvement in data quality of B0-sensitive MRI methods could be achieved.
Introduction
A spatially homogeneous static magnetic field (B0) is a prerequisite for several MRI methods. A change in the patient’s position causes motions artefact and decreases the homogeneity of the B0 field1–3. Changes in B0 are increasingly problematic at ultra-high-field MR scanners4. Several internal (e.g. navigators5) and external tracking methods (e.g. optical tracking6) or changes of the B0 field (e.g. NMR probes7) have been proposed. We propose a deep learning approach to map the change of the B0 field within the brain from the change of the head position. An optimized U-net is used to predict B0 maps.Methods
A 7T Magnetom+ MR Scanner (Siemens Healthineers, Erlangen, Germany) with a 32-channel head coil (Nova Medical) was used to scan 13 healthy volunteers (8 males and 5 females). For each volunteer, an anatomical reference (MP2RAGE sequence8) and B0 maps at 30 head positions (double-echo gradient echo (GRE)-MRI sequence) were measured. For the B0 maps scans, the volunteers were asked to change head position randomly within a comfortable range. MP2RAGE datasets were down-sampled to the resolution of B0 maps. MP2RAGE data were indirectly co-registered (Flirt, FSL toolbox9) by applying transformation matrices from the co-registration of the magnitude images of the first echo (of the GRE-sequence) between the first position and the rest of the positions.The training dataset consists of the data of 8 volunteers (7 males and 1 female). Each instance contains the input: (i) anatomical MRI (T1-weighted MP2RAGE) at the initial position, (ii) B0 map at the initial position, and (iii) the same anatomical MRI, but after applying the 6DoF transformation (new position); and the output (i) B0 map at the new position. All training datasets were augmented by adding randomly scaled spherical harmonics B0 fields. The testing dataset consists of the data of 5 additional volunteers. The same pre-processing as for the training data was done but without augmentation.
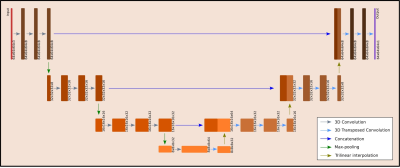
All calculations were performed on a DGX station equipped with four Tesla V100 GPU cards (Nvidia, Santa Clara, CA, US). The PyTorch10 and the Neural network intelligence (NNI)11 were used. The 3D U-net architecture (Figure 1) was parameterized. The Adam optimizer was used. Learning rate, mini-batch size and the parameters of the network architecture were optimized by the NNI. The mean-squared error was used. The NNI optimization was run over 200 trials for 50 epochs per trial. The final training with the optimized parameters was performed using 1500 epochs.
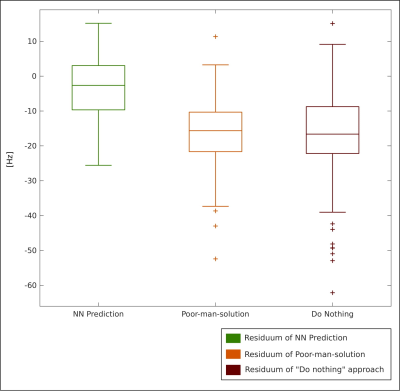
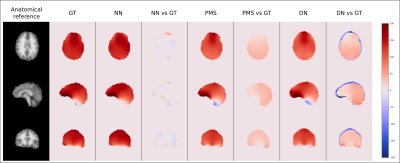
The accuracy of the neural network (NN) to predict B0 maps was compared with two approaches: (i) to a “poor-man-solution” (PMS), for which the B0 map at the initial head position was simply co-registered to the new position and (ii) to a “do-nothing” (DN) approach, for which the B0 map was not updated for the new position at all. The approaches were compared against the ground truth B0 maps (GT). The overall performance of the methods was compared as boxplots of the medians of residua of the methods for all volunteers’ positions in testing data. The quality of the NN B0 maps was compared relative to the maps of the other two methods and their residua maps to the GT B0 maps.
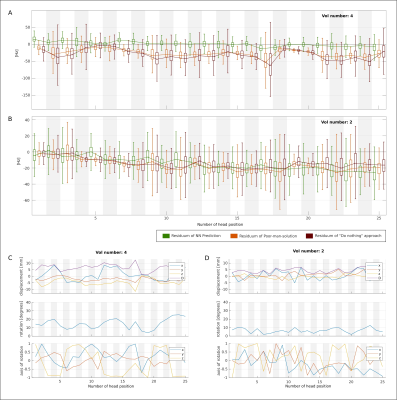
A quantitative evaluation of each volunteer and for each head position was conducted. The boxplots of the residua were compared between the methods and the percentage of the head positions was derived, in which the NN-predicted B0 maps outperformed the other approaches.
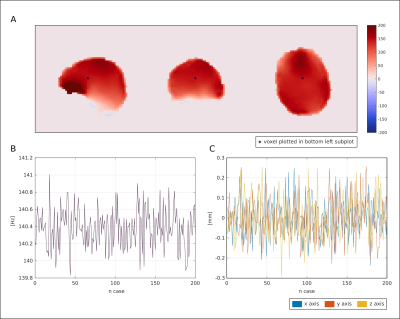
The precision of the network prediction was tested by simulating a small movement of a patient (translations in the spatial directions parametrized by a Gaussian distribution (mean=0, stddev=0.1mm)), which is comparable to uncertainties of navigators3.
Results
The results of the neural architecture search are depicted in Figure 1. Boxplots of overall performance for the NN prediction, the PMS and the DN approach compared to the GT are plotted in Figure 2. The B0 maps of a representative volunteer of all approaches together with the ground truth and the anatomical reference are depicted in Figure 3. Overall in four cases out of five, the NN outperformed the PMS and the DN. However, the results of volunteer #2 diverged from the rest. Quantitative results of volunteer #4 and volunteer #2 are in Figure 4. The results of the precision analysis are in Figure 5.Discussion and Conclusion
This abstract presents the results for a new deep learning-based approach to predict B0 maps based on patient movement. The prediction of the B0 maps was carried out by a U-net neural network, trained with the experimentally acquired data of eight volunteers. The performance of the network was compared with two other approaches using a test dataset of five additional volunteers. The estimated B0 maps of all approaches were compared against the ground truth and the stability of the network prediction was tested. The results suggest, that the prediction of B0 maps is feasible and by combination with external tracking a considerable improvement in data quality of B0-sensitive MRI methods could be achieved.Acknowledgements
FWF grant number: P 34198References
1. Bogner W, Hess AT, Gagoski B, et al. Real-time motion- and B0-correction for LASER-localized spiral-accelerated 3D-MRSI of the brain at 3T. Neuroimage 2014;88:22–31 doi: 10.1016/J.NEUROIMAGE.2013.09.034.
2. Liu J, de Zwart JA, van Gelderen P, Murphy-Boesch J, Duyn JH. Effect of head motion on MRI B 0 field distribution. Magn. Reson. Med. 2018;80:2538–2548 doi: 10.1002/MRM.27339.
3. Heckova E, Považan M, Strasser B, et al. Real-time Correction of Motion and Imager Instability Artifacts during 3D γ-Aminobutyric Acid-edited MR Spectroscopic Imaging. Radiology 2018;286:666–675 doi: 10.1148/RADIOL.2017170744.
4. Juchem C, de Graaf RA. B0 magnetic field homogeneity and shimming for in vivo magnetic resonance spectroscopy. Anal. Biochem. 2017;529:17–29 doi: 10.1016/J.AB.2016.06.003.
5. White N, Roddey C, Shankaranarayanan A, et al. PROMO: Real-time prospective motion correction in MRI using image-based tracking. Magn. Reson. Med. 2010;63:91–105 doi: 10.1002/MRM.22176.
6. Zaitsev M, Dold C, Sakas G, Hennig J, Speck O. Magnetic resonance imaging of freely moving objects: prospective real-time motion correction using an external optical motion tracking system. Neuroimage 2006;31:1038–1050 doi: 10.1016/J.NEUROIMAGE.2006.01.039.
7. Zanche N De, Barmet C, Nordmeyer-Massner JA, Pruessmann KP. NMR probes for measuring magnetic fields and field dynamics in MR systems. Magn. Reson. Med. 2008;60:176–186 doi: 10.1002/MRM.21624.
8. Marques JP, Gruetter R. New developments and applications of the MP2RAGE sequence--focusing the contrast and high spatial resolution R1 mapping. PLoS One 2013;8:e69294 doi: 10.1371/journal.pone.0069294.
9. Jenkinson M, Bannister P, Brady M, Smith S. Improved Optimization for the Robust and Accurate Linear Registration and Motion Correction of Brain Images. 2002 doi: 10.1006/nimg.2002.1132.
10. Paszke A, Gross S, Chintala S, et al. Automatic differentiation in PyTorch. In: NIPS-W. ; 2017.
11. GitHub - microsoft/nni: An open source AutoML toolkit for automate machine learning lifecycle, including feature engineering, neural architecture search, model compression and hyper-parameter tuning. https://github.com/microsoft/nni. Accessed September 10, 2021.
Figures