0946
Deep Learning based MR reconstruction for accelerated 3D-PREFUL ventilation assessment of post-COVID-19 patients from undersampled MR-images1Institute of Diagnostic and Interventional Radiology, Hannover Medical School, Hannover, Germany, 2Biomedical Research in Endstage and Obstructive Lung Disease Hannover (BREATH), German Center for Lung Research (DZL), Hannover, Germany
Synopsis
3D phase-resolved functional lung (3D-PREFUL) proton MRI enables a radiation-free and non-contrast-enhanced ventilation assessment of human lungs. However, generating high-quality images usually requires a long acquisition time. Acceleration can be achieved by undersampling k-space data, but the resulting violation of the Nyquist theorem leads to image artifacts. Deep learning (DL)-based reconstruction approaches are proposed as a solution for this dilemma. Two novel loss functions are introduced to create a deep learning based reconstruction, optimized for lung MRI. The feasibility of ventilation assessment, including ventilation defect identification, from 8x undersampled MR-images of post-COVID-19 patients, reconstructed by a neural network is demonstrated.
Introduction
3D phase-resolved functional lung (3D-PREFUL)-MRI1,2 enables non-contrast-enhanced ventilation assessment of human lungs. However, the generation of high-quality images usually requires a long acquisition time. A reduction of acquisition time can be achieved by k-space undersampling, but the violation of the Nyquist theorem leads to image artifacts. In addition to compressed sensing, deep learning (DL)-based reconstruction approaches3,4 have been suggested as a solution for this dilemma. Qin et al.3 published a convolutional, recurrent neural network architecture (CRNN) for the data-consistent image reconstruction from undersampled, cardiac cine-MRI-scans. We reused the provided network architecture to examine the feasibility of reconstructing the ventilation signal of post-COVID-19 patients from undersampled 3D-PREFUL-MR-images using recurrent neural networks. In addition, two novel loss functions tailored for lung-MRI are introduced.Methods
22 healthy volunteers (age range: 18-65, 9 male) and 21 post-COVID-19 patients (age range: 23-71,13 male) underwent imaging on a 1.5T MR-scanner (MAGNETOM Avanto, Siemens Healthcare, Erlangen, Germany). Following sequence parameters were used for 3D-PREFUL-MRI2: FOV 50×50cm2, matrix size 128×128, slice thickness 4mm, TE 0.81ms, TR 1.9ms, flip angle 3.5°, and pixel bandwidth 1500Hz/pixel.Since the CRNN requires a uniform data shape, the 3D-PREFUL-MR-scans consisting of 52-80 slices with 30-43 respiratory phases were harmonized to overlapping sequences of 30 respiratory phases and combined to a dataset of 5088 30x128x128 records. As shown in Figure 1, the dataset was divided into a training set (70%) and a test set (30%), the training set was divided again into 70% final training data and 30% validation data.
The CRNN was implemented in Pytorch 1.7. Overall, 18 different versions were trained over 25 epochs on a NVIDIA-DGX-A100 GPU. As shown in Figure 2a-c, image series were artificially undersampled using Cartesian undersampling masks described by Qin et al.3. The random removal of k-space lines was repeated in each epoch, resulting in 25 different undersampled versions for each image.
An initial CRNN was trained to minimize a loss calculated by the mean squared error (MSE) between the original image (Figure 2a) and the predicted image (Figure 2d). As an alternative to the image-based MSE, called global-image-loss (GL), two novel MSE-based loss functions tailored for DL-based reconstruction of 3D-PREFUL-MRI-data were introduced: Thoraxquad-loss (TQ) only comprises reconstruction errors within a rectangular region of interest (ROI) enclosing heart and lung. 2-quads-loss (2Q) multiplies the errors within the same rectangle with 0.5 and errors from an additional rectangle surrounding the diaphragm with 0.25, the sum of both errors defines the 2Q-loss. The concept of both loss functions is visualized in Figure 3.
Initially, 9 different CRNNs were trained using the three loss functions for 4x, 8x and 12x undersampled MR-data.
Additionally, further 9 CRNNs were solely trained with data from healthy volunteers, to examine the impact on the reconstruction of post-COVID-19 lungs using a deep learning model that only knows healthy lungs.
The reconstruction results of overall 18 different CRNNs were compared by calculating MSE as well as peak-signal-to-noise-ratio (PSNR) between fully sampled ground truth and images reconstructed by a CRNN. Additionally to the GL-loss, also reconstruction errors within the ROI of the TQ-loss function were compared.
Finally, 8x undersampling was chosen as the best trade-off between undersampling level and image quality to evaluate the feasibility of conducting 3D-PREFUL ventilation analysis of post-COVID-19 patients from undersampled MRI-data. Regional ventilation (RVent) map and ventilation defect percentage (VDP) map were computed1 for reconstructions results from CRNNs trained with GL-, TQ- and 2Q-loss function. The three different RVent-maps were compared to the ground truth by structural similarity index (SSIM), the corresponding VDP-maps were compared by dice coefficient to defects from VDP-Maps of the fully sampled scan.
Results
The introduced loss functions decrease the error in DL-based reconstruction especially from 8x and 12x undersampled MR-images of post-COVID-19 patients.Figure 4 shows that TQ-loss produces the highest SSIM at RVent-maps, highest Dice coefficient at VDP-maps and results in a difference of 2 percent VDP in comparison to the fully sampled data.
As summarized in Table 1, CRNNs trained with data from healthy volunteers only did not significantly increased the reconstruction error.
A PSNR improvement of 9dB for 4x, 8dB for 8x and 5dB for 12x undersampled data was measured.
Discussion
CRNNs can reconstruct remarkable results from highly undersampled lung MR-images and facilitate 3D-PREFUL ventilation assessment. However, two effects could be observed:The CRNN architecture is better in predicting anatomy than ventilation signal. Integrating the introduced loss functions can correct this behavior; Replacing the rectangular ROIs by more precise segmentation masks could enforce the correction effect.
Secondly, the feasibility of reconstructing pulmonary vessels seems to be limited. Vessel connections were blurry or missing in images having a huge reconstruction error. Considering the image quality of massively undersampled images, a comparison with other reconstruction approaches is necessary to determine whether reconstruction of fine vessel structures from such data is possible at all.
Conclusion
3D-PREFUL of highly undersampled MRI-data from post-COVID-19 patients seems to be feasible. Loss functions tailored for lung reconstruction leads to DL-models with improved reconstruction accuracy of anatomy and signal, crucial for 3D-PREFUL.Furthermore, DL-based MR-reconstruction tools for COVID-19 diagnostics can be trained with data from healthy volunteers only, as this does not increase the error significantly.
Acknowledgements
This work was supported by the German Centre for Lung Research (DZL). The authors thank Melanie Pfeifer and Frank Schröder for their help during data acquisition.References
- Klimeš, F., Voskrebenzev, A., Gutberlet, M., et al. 3D phase‐resolved functional lung ventilation MR imaging in healthy volunteers and patients with chronic pulmonary disease. Magnetic Resonance in Medicine. 2021;, 85(2), 912-925. doi: 10.1002/mrm.28482
- Klimeš, F., Voskrebenzev, A., Gutberlet, M., et al. Repeatability of dynamic 3D phase-resolved functional lung (PREFUL) ventilation MR Imaging in patients with chronic obstructive pulmonary disease and healthy volunteers. Magnetic Resonance in Medicine. 2021; 54(2),618-629. doi: 10.1002/jmri.27543
- Qin, C., Schlemper, J., Caballero, J., Price, A. N., et al. Convolutional Recurrent Neural Networks for Dynamic MR Image Reconstruction. IEEE Transactions on Medical Imaging. 2018; 38(1), 280-290. doi: 10.1109/TMI.2018.2863670
- Ramzi, Z., Ciuciu, P., & Starck, J. L. . Benchmarking MRI reconstruction neural networks on large public datasets. Applied Sciences. 2020; 10(5), 1816. doi: 10.3390/app10051816
- Uecker M, Ong F, Tamir JI, et al. Berkeley advanced reconstruction toolbox. Proc Intl Soc Mag Reson Med. 2015;23:2486. doi:10.1002/mrm.25176
Figures

Figure 1: Balanced dataset consisting of 22 healthy volunteers and 21 post-COVID-19 patients, which resulted in final dataset size of 5088 image sequencs of 30 respiratory phases and image matrix of 128x128. Training set contained 70% and test set contained 30% of study population data.
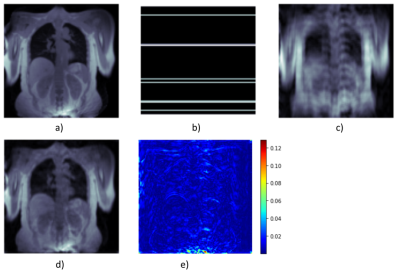
Figure 2: Fully sampled image reconstructed using parallel imaging with iterative compressed sensing reconstruction implemented in Berkeley Advanced Reconstruction Toolbox5 (a) is artificially undersampled by a Cartesian mask (b). DL-based reconstruction transforms undersampled image (c) to (d) by considering the undersampling maks as well as the undersampled k-space. (e) shows the difference image between (a) and (d).
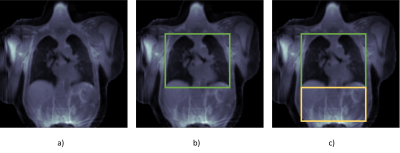
Figure 3: Three different loss functions to be minimized during the training process of a neural network: (a) Global image loss: aims to minimize the reconstruction error of the whole image (b) TQ-loss: reconstruction errors within the green rectangle increases the loss much more than errors in other image regions c) 2Q-loss: errors in reconstruction of the diaphragm increase the loss much more than errors in other regions, errors in reconstructing the lung parenchyma and heart increase the loss more than errors in reconstructing the diaphragm.
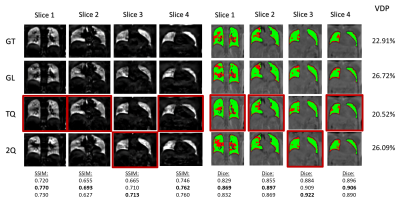
Figure 4: Comparison of 3D-PREFUL derived RVent maps (on the left) and corresponding VDP-maps (on the right) resulting from three differently trained CRNNs . Red borders mark the reconstruction with the lowest difference to the ground truth (GT).
The network trained with the TQ-loss function (TQ) outperformed the network trained with a GL-loss function (GL) and performs better than the 2Q-loss function (2Q) in 3 of 4 slices. Overall, images reconstructed by that network results in a VDP value with the smallest difference (2%) to the VDP value of the fully sampled scan of the same patient.