0945
MRI Regularization for Low-Count PET Image Recovery using Unsupervised Learning Without the Need of Training Set - A Multi-Tracer PET/MRI Study1Biomedical Engineering, Stony Brook University, Stony Brook, NY, United States, 2Radiology, Stony Brook Medicine, Stony Brook, NY, United States, 3Psychiatry, Stony Brook Medicine, Stony Brook, NY, United States
Synopsis
Unsupervised denoising is useful as it allows low-count PET image recovery without the need of paired training data(low-count/full-count). However, current unsupervised denoising models utilize Contrast-to-Noise Ratio as stopping criteria to optimize the image recovery process, which can be improved by considering structural information to maintain the integrity of gross anatomy. In this work, we proposed an MRI structural regularization loss function for low-count PET image recovery using an unsupervised learning model, which does not require paired training sets and demonstrated that the proposed method is superior in both qualitative and quantitative analyses for two radiotracers with very different physiological uptake.
Introduction
Deep learning techniques have been used to recover high quality image from low-dose PET images. Recent works have utilized MRI as an anatomical prior in deep learning to provide better PET denoising results (1). While previous deep learning techniques for PET denoising have used supervised learning techniques, which require image pairs of low-count and paired high quality PET image, it is difficult to accumulate large, paired data to train such a network, particularly for less common tracers. These intrinsic difficulties for training supervised learning models make it difficult to implement for newly developed tracers that have limited datasets.More recently, an unsupervised learning technique has used MRI as deep image prior for PET denoising. This networks incorporates a U-Net based deep learning framework(2) with no need for pairs of low-count and high-count images. Without having full-count PET data to train the network, the study utilized contrast to noise ratio as a stopping criterion (CNRstop) to optimize the denoising process. CNR, although useful to optimize, requires segmentation of regions to calculate and does not consider the overall structural anatomy within the loss function (3). In this work we introduce a novel regularization loss function for unsupervised PET denoising that considers structural MRI to recover for 10% reduced low-count PET. Moreover, this regularized loss function is examined across multiple tracers including 18F-FDG and 18F-VAT.
Methods
Data was collected across two different PET radiotracers. 111-185 MBq (3-5mCi) of 18F-FDG or 18F-VAT was administered. This study acquired listmode data using a dedicated MRI head coil for a Siemens Biograph mMR PET/MRI scanner for three 18F-FDG datasets and three 18F-VAT PET datasets. Attenuation maps were generated using an established MRI-based algorithm(4, 5). Hardware attenuation maps were also extracted for reconstruction. Low-count PET data (10% count) were generated through Poisson thinning from the listmode file. Single static 18F-FDG and 18F-VAT PET images were reconstructed from 10-minute emission data (50-60 minutes after injection) and 60-minute emission data (90-150 minutes after injection), respectively, using Siemens’ E7tools with ordered subset expectation maximization (OSEM). The 3D U-Net architecture was used for the unsupervised learning network as previously employed (Figure 1)(3). Similar to previous unsupervised learning framework(6), anatomical MPRAGE T1 weighted images were acquired with TR=2300ms, TE=3.24ms, flip angle 9º, voxel size = 0.66mm. T1 weighted image was used as input to the network and were trained to output low-count PET image; therefore, not relying on training pairs with full-count PET.This network used an L2 loss function with respect to low-count PET. However, the loss function for this work also utilized an MRI structural regularization that incorporated structural similarity metric (SSIM); wherein SSIM was calculated between network output and structural MRI that was used as input.
LMRI_reg = (1-λ)L2 + λ (Σni=1 SSIMi)/n
Here, SSIM was calculated times, each with different size gaussian filters. The loss function was calculated with n=4 and λ =0.25. For comparison, we used the previously utilized stopping criteria of CNR. Final outputs from the unsupervised learning U-Net framework were then compared to full-count data to assess whether the MRI structural regularization loss function outperformed the previously used method of CNR as stopping criteria. Evaluation of network output and full-count images was completed using: 1) Peak Signal to Noise Ratio (PSNR), 2) Structural Similarity (SSIM), 3) Root Mean Square Error (RMSE).
Results
Figure 2 shows network output for our proposed method and the previously used CNRstop method. Table 1 also provides quantitative analysis across methods, wherein our proposed method outperformed the CNRstop across all quantitative metrics and across both radiotracers.Discussion
In this work we have developed a regularization loss function for unsupervised denoising that considered structural MRI to recover 10% low-count PET images across multiple tracers: 18F-FDG and 18F-VAT. This is the first study to develop a regularized loss function for unsupervised denoising and evaluated across multiple radiotracers. Our proposed method was compared to the previously developed CNRstop, which although recovers PET images from low-count, does not consider the overall structural integrity of the PET image. Visual assessment of CNRstop vs our proposed method shows that regularized loss function provides better structural integrity in the striatum across 18F-FDG and thalamus across 18F-VAT. Considering that 18F-VAT has very high uptake in the striatum and 18F-FDG has more uniform uptake across the brain, the proposed regularized loss function shows to generalize across both tracers very well indicating that it is not tracer dependent like most supervised learning networks.Conclusion
Previous unsupervised PET denoising frameworks have used a U-Net architecture and CNR either in a lesion or healthy tissue as stopping criteria. We employed an MRI structural regularization loss function that produced better results for multiple radiotracers, highlighting its reproducibility and feasibility across institutions that do not have large dataset resources.Acknowledgements
This work is in part supported by the Parkinson's Foundation.References
1. Onishi Y, Hashimoto F, Ote K, Ohba H, Ota R, Yoshikawa E, Ouchi Y. Anatomical-guided attention enhances unsupervised PET image denoising performance. Medical Image Analysis. 2021;74:102226.
2. Lempitsky V, Vedaldi A, Ulyanov D, editors. Deep image prior. 2018 IEEE/CVF Conference on Computer Vision and Pattern Recognition; 2018: IEEE.
3. Cui J, Gong K, Guo N, Wu C, Meng X, Kim K, Zheng K, Wu Z, Fu L, Xu B. PET image denoising using unsupervised deep learning. European journal of nuclear medicine and molecular imaging. 2019;46(13):2780-9.
4. Chen KT, Izquierdo-Garcia D, Poynton CB, Chonde DB, Catana C. On the accuracy and reproducibility of a novel probabilistic atlas-based generation for calculation of head attenuation maps on integrated PET/MR scanners. European journal of nuclear medicine and molecular imaging. 2017;44(3):398-407. PubMed PMID: 27573639 PMCID: 5285302
5. Poynton CB, Chen KT, Chonde DB, Izquierdo-Garcia D, Gollub RL, Gerstner ER, Batchelor TT, Catana C. Probabilistic atlas-based segmentation of combined T1-weighted and DUTE MRI for calculation of head attenuation maps in integrated PET/MRI scanners. American journal of nuclear medicine and molecular imaging. 2014;4(2):160. PubMed PMID: 24753982.
6. Cui J, Gong K, Guo N, Wu C, Meng X, Kim K, Zheng K, Wu Z, Fu L, Xu B, Zhu Z, Tian J, Liu H, Li Q. PET image denoising using unsupervised deep learning. European Journal of Nuclear Medicine and Molecular Imaging. 2019;46(13):2780-9. doi: 10.1007/s00259-019-04468-4.Figures
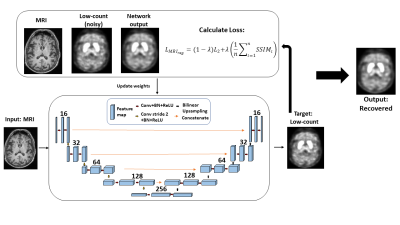
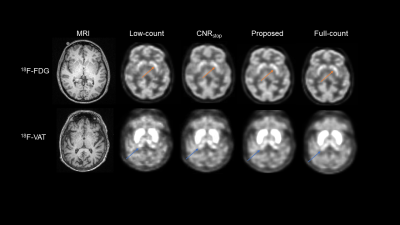
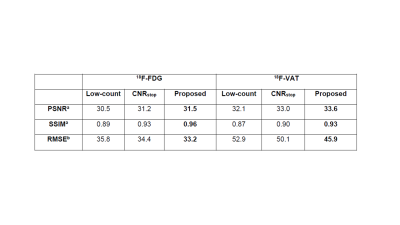
Table 1: Quantitative metrics evaluated between low-count, the previously used CNR as stopping criteria and our proposed method all relative to full-count images.
a Higher is better
b Lower is better
PSNR: Peak Signal to Noise Ratio
SSIM: Structural Similarity Index
RMSE: Root Mean Square Error