0944
Gradual RAKI reconstruction merged with intermittent GRAPPA blurring moderation with limited scan-specific training samples1Brain Imaging Centre, Research Centre for Natural Sciences, Budapest, Hungary, 2Department of Physics, University of Würzburg, Würzburg, Germany, 3Magnetic Resonance and X-Ray Imaging Department, Development Center X-ray Technology EZRT, Fraunhofer Institute for Integrated Circuits IIS, Würzburg, Germany
Synopsis
RAKI is a scan-specific k-space interpolation technique based on deep convolutional networks, which bears superior noise resilience compared to GRAPPA. However, RAKI may introduce severe blurring in image reconstruction due to reduced number of autocalibration signal lines at higher acceleration factors. We propose Gradual RAKI, which exhibits the benefit of mixing RAKI and GRAPPA in a preparatory block for data augmentation purposes prior to a conventional RAKI reconstruction. Data augmentation provides an effective way to create synthetic ACS lines out of 8 original ACS lines at 4-fold acceleration, while valuable features are retained from both RAKI and GRAPPA reconstruction methods.
Introduction
RAKI [1] is a scan-specific k-space interpolation technique based on deep convolutional networks. RAKI bears superior noise resilience compared to GRAPPA [2], resulting in improved image quality. However, RAKI may introduce severe residual artefacts and blurring in conventional in-plane image reconstruction due to reduced number of autocalibration signal (ACS) lines at higher acceleration factors. In this work, we propose Gradual RAKI, where k-space segments are sequentially filled up with the interpolated data by subsequent RAKI reconstructions intermitted with a single GRAPPA reconstruction to mitigate RAKI-introduced blurring. This work has been conceived due to fruitful discussions on Iterative RAKI, which has been presented at the annual meeting of the ESMRMB 2021 congress [3].Methods
Gradual RAKI consists of a few consecutive reconstruction steps only, that can be divided into a preparatory block and a robust RAKI block (Fig. 1). The former aims to generate synthetic ACS lines that can be fed to the latter as the final reconstruction step. The preparatory components of Gradual RAKI’s pipeline serve two distinct purposes, and thus are divided into two sections. First, two consecutive RAKI reconstructions are carried out to serve as initial data augmentation steps (i.e., synthetic ACS line generation). Due to its superior noise resilience compared to GRAPPA, RAKI is assigned as a preferable method for initial data augmentation to minimize noise propagation into the synthetic ACS data set. Second, beyond further increasing the synthetic ACS lines, a single GRAPPA reconstruction is performed to counterbalance the potentially RAKI-introduced residual artefacts and blurring, stemming from the low number of original ACS lines. As the final step of Gradual RAKI, a RAKI reconstruction synthesizes the missing lines of the whole accelerated measurement bearing a sufficient number of synthetic ACS lines originating from the preparatory block.Rather than opting for a single robust neural network with sufficient number of ACS lines to adequately generalize for all frequency components, we designed Gradual RAKI’s consecutive reconstruction steps to remain within the range of their generalization capacity based on the extent of available k-space training data.
Each reconstruction output serves as training input for the consecutive step. Original ACS lines are reinserted after every reconstruction step. 4,6,8 ACS lines are assigned as excessively limited original training data for acceleration factor R = 2,3,4, respectively, i.e., 2 x acceleration factor as ACS lines. In the data augmentation phase of the preparatory block, the output size is selected to be 2 x acceleration factor greater than input size, thus the undersampled k-space is filled up symmetrically with respect to k-space origin. Complex-valued convolution [4][3] is implemented to involve correlation between real and imaginary parts representing k-space phase information. Complex ReLU [5] is chosen as activation and mean-squared-error (MSE) as loss function. The number of epochs is set to 300, with MSE normalized to the first iteration error dropping to 1e-4 as the early stopping condition. Learning rate is set to 3e-3 universally and training weights are randomly initialized in every case as all reconstruction steps are treated individually.
2D in vivo brain data were collected for evaluation with a 32-channel receiver coil on a clinical scanner on a healthy volunteer at 3 T (Magnetom Prisma, Siemens Healthineers, Erlangen, Germany). One set (ref. to as neuro_T1) was acquired using TSE with TR/TE = 350/2.46 ms, FOV = 220 x 193 mm2, matrix size 256 x 224. Another set (ref. to as neuro_T2) was acquired using Inversion recovery TSE (FLAIR) with TR/TE = 9000/111 ms, FOV = 220 x 193 mm2, matrix size 256 x 224.
Quantitative visual remarks and difference images are displayed with respect to the fully sampled reference data set. Normalized Mean Squared Error (NMSE), Peak Signal to Noise Ratio (PSNR) and Structural Similarity Index Metric (SSIM) computed with respect to the fully sampled reference data set serve as image quality metrics. GRAPPA and RAKI reconstructions are carried out from the original ACS lines to serve as comparison baseline.
Results
Gradual RAKI shows improved image quality compared to RAKI and GRAPPA at 4-fold acceleration even with 8 ACS lines as excessively limited original training data. (Fig. 2 and Fig. 3). Image quality metrics demonstrate the performance of Gradual RAKI compared to GRAPPA and RAKI at 2,3,4-fold accelerations with the application of 4,6,8 ACS lines, respectively. (Fig. 4).Discussion
Gradual RAKI exhibits the benefit of mixing RAKI and GRAPPA in a preparatory block for data augmentation purposes prior to a conventional RAKI reconstruction. Data augmentation provides an effective way to create synthetic ACS lines out of 8 ACS lines at 4-fold acceleration, while valuable features are retained from both RAKI and GRAPPA reconstruction methods.In the original RAKI article of Akçakaya et al. [1], GRAPPA was proposed to generate synthetic ACS lines. However, in Gradual RAKI’s approach, an initial RAKI reconstruction was chosen for data augmentation purposes in the preparatory block and GRAPPA was only used as an intermittent step to mitigate RAKI-related artefacts.
Acknowledgements
This work has been supported by grants from the Hungarian Brain Research Program (Nemzeti Agykutatási Program 2017-1.2.1-NKP-2017-00002).
References
[1] Akcakaya M, Moeller S, Weingartner S, Uğurbil K. Scan-specific robust artificial-neural-networks for k-space interpolation (RAKI) reconstruction: Database-free deep learning for fast imaging. Magnetic Resonance in Medicine 2019;81(1):439–453.
https://onlinelibrary.wiley.com/doi/abs/10.1002/mrm.27420.
[2] Griswold MA, Jakob PM, Heidemann RM, Nittka M, Jellus V,Wang J, et al. Generalized autocalibrating partially parallel acquisitions (GRAPPA). Magnetic Resonance in Medicine 2002;47(6):1202–1210.
https://onlinelibrary.wiley.com/doi/abs/10.1002/mrm.10171.
[3] Dawood P, Blaimer M, Breuer F, Burd PR, Jakob PM, Oberberger J. Iterative RAKI with complex-valued convolution for improved image reconstruction with limited scan-specific training samples. In: Magnetic Resonance Materials in Physics, Biology and Medicine, vol. 34; 2021. p. S24–S25.
https://doi.org/10.1007/s10334-021-00947-8.
[4] Cole E, Cheng J, Pauly J, Vasanawala S. Analysis of deep complex-valued convolutional neural networks for MRI reconstruction and phase-focused applications. Magnetic Resonance in Medicine 2021;86(2):1093–1109.
https://onlinelibrary.wiley.com/doi/abs/10.1002/mrm.28733.
[5] Trabelsi C, Bilaniuk O, Zhang Y, Serdyuk D, Subramanian S, Santos JF, et al. Deep Complex Networks. In: International Conference on Learning Representations; 2018.
Figures
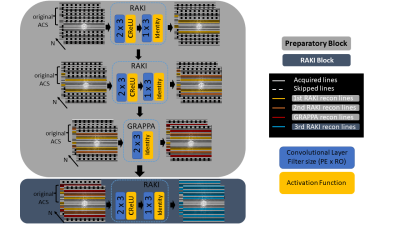
Fig. 1: Workflow of Gradual RAKI. The input layer for the preparatory block of RAKI is assigned 2 x 3 kernel size (PE x RO), equal to the one of GRAPPA. CRelu is chosen as activation function between the input and output layers. No hidden layers are built in the design due to the low number of degrees of freedom provided by the extremely limited number of original ACS lines. PE: phase-encoding direction; RO: read-out direction; CRelu: Complex Rectified Linear Unit
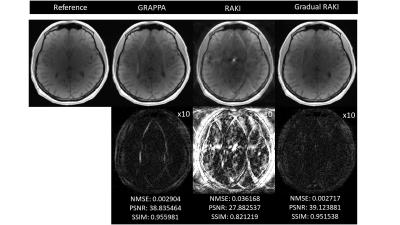
Fig. 2: GRAPPA (left), RAKI (center) and Gradual RAKI (right) reconstructed images in comparison for neuro_T1 data set for R = 4 with ACS = 8 as original training data set (top). Difference images with respect to the fully sampled reference image are displayed with 10x scaling for better visibility (middle). Normalized Mean Squared Error (NMSE), Peak Signal to Noise Ratio (PSNR) and Structural Similarity Index Metric (SSIM) serve as quantitative metrics with respect to the reference image (bottom).
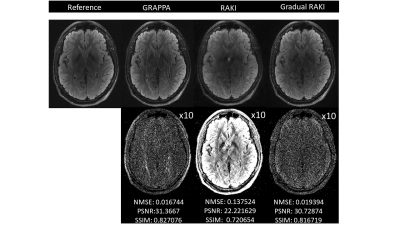
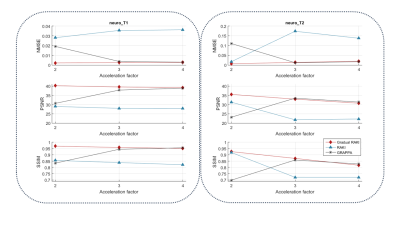
Fig. 4: Normalized Mean Squared Error (NMSE, top row), Peak Signal to Noise Ratio (PSNR, middle row) and Structural Similarity Index Metric (SSIM, bottom row) are computed with respect to the fully sampled reference image for 2,3,4-fold in-plane acceleration with ACS = 4,6,8 original training lines, respectively, for both neuro_T1 (left column) and neuro_T2 (right column) in vivo data sets. Gradual RAKI exhibits improved performance for neuro_T1 data set in all metrics, whereas performance upgrade is more moderate for neuro_T2 data set.