0925
Automatic Prostate Tumor Segmentation: Does the Convolutional Neural Network Learn How the Tumor Looks, or What the Radiologist Sees?
Deepa Darshini Gunashekar1, Lars Bielak1,2, Benedict Oerther 3, Matthias Benndorf 3, Anca Grosu 2,3, Constantinos Zamboglou2,3, and Michael Bock 1,2
1Dept.of Radiology, Medical Physics, Medical Center University of Freiburg, Faculty of Medicine, University of Freiburg, Freiburg, Germany, 2German Cancer Consortium (DKTK), Partner Site Freiburg, Freiburg, Germany, Freiburg im Breisgau, Germany, 3Dept.of Radiology, Medical Center University of Freiburg, Faculty of Medicine, University of Freiburg, Freiburg, Germany
1Dept.of Radiology, Medical Physics, Medical Center University of Freiburg, Faculty of Medicine, University of Freiburg, Freiburg, Germany, 2German Cancer Consortium (DKTK), Partner Site Freiburg, Freiburg, Germany, Freiburg im Breisgau, Germany, 3Dept.of Radiology, Medical Center University of Freiburg, Faculty of Medicine, University of Freiburg, Freiburg, Germany
Synopsis
A convolutional neural network was implemented to automatically segment tumors in multi-parametric MRI data. The influence of the variability in the ground truth data was evaluated for automated prostate tumor segmentation. Therefore, the agreement between the predictions of the CNN was measured with co-registered whole mount histopathology images and the tumor contours drawn by an expert radio-oncologist. The results indicate that the network can discriminate tumor from healthy tissue rather than mimicking the radiologist.
Introduction
Multiparametric magnetic resonance imaging (mpMRI) is currently used in clinical practice as the standard protocol for diagnosing, staging, and definitive management of prostate carcinoma (PCa)1. mpMRI demonstrated excellent sensitivity in the detection of PCa by providing high soft-tissue contrast and better differentiation of internal structures and surrounding tissues of the prostate gland (PG). The segmentation of prostate substructures in mpMRI is the basis for clinical decision-making. Due to the complexity associated with the location and size of the PG, an accurate manual delineation of PCa is time consuming and susceptible to high inter- and intra-observer variability 2. Algorithms based on convolutional neural networks (CNNs) have shown promising results for PCa segmentation 3-7.Even though CNNs have shown to perform well in tumor segmentation, it is difficult to quantify their segmentation quality in the absence of verified ground truth data. Whole mount histopathology of prostate cross sections from patients who underwent radical prostectomy have been used to study the correlation of mpMRI for definition of intraprostatic tumor mass 8-10.This data can be used as true ground truth in evaluating the CNNs.To evaluate the quality of the CNN predictions, we compare the agreement between the CNN predicted PCa segmentations with PCa contours transferred from whole mount histopathology slices that are co-registered with mpMRI images, and PCa contours drawn by expert radiooncologist on mpMRI data.Materials and Methods
mpMRI data from PCa patients was used that were acquired between 2008 and 2019 on clinical 1.5T and 3T systems. Patient data was separated into two groups: (a) a large irradiation and prostatectomy group (n= 122), and (b) a prostatectomy group (n = 15) from which whole organ histopathology slices were available. For CNN training only data from group (a) was used, which contained pre-contrast T2-weighted TSE and apparent diffusion coefficient (ADC) maps together with computed high b-value maps (b = 1400 s/mm²). In the images the entire PG and the PCa (Gross Tumor Volume, GTV-Rad) were contoured by a radiation oncologist with over 5 years of experience, in accordance to the PI-RADs v2 standards11.The histology based GTV-Histo contours were generated as in12. Image data were cropped to a smaller FOV around the PG and then registered and interpolated to an in-plane resolution of 0.78×0.78x3 mm³. A patch-based 3D CNN of the UNET architecture13 was trained for the automatic segmentation of GTV and PG. The network was implemented in MATLAB® (2020a,Math Works,Inc.,Natick/MA). The optimal parameter set was obtained by a Bayesian optimization scheme to maximize the segmentation performance and used to train the network within 150 epochs on an NVIDIA RTX2080 GPU. The center location of each patch was chosen randomly with respect to the original image. The probability of the center pixel to be of the class background, GTV or PG was set to 33% to account for class imbalance and a chance of 70% for a random 2D-rotation in the axial plane was added for data augmentation. During the CNN testing phase, the mpMRI data from the prostatectomy group14 was used as a test cohort to evaluate the quality of the networks prediction for PCa segmentation. The resulting segmentation was then compared via the Dice Sorensen Coefficient (DSC) with the ground truth from GTV–Histo (Histo) and GTV-Rad (Rad).Results
Figure 1 shows the ground truth and the predicted segmentations overlaid on the corresponding input image sequences for 3 test patients from the test cohort.The mean, standard deviation and median DSC between the CNN-predicted segmentation and the ground truth across the test cohort was 0.31,0.21 & 0.37 (range: 0 - 0.64) for CNN-Histo and 0.32,0.20 &0.33 (range: 0 - 0.80) for CNN-Rad, respectively (Fig.2). A two-sided students t-test on the segmentation performance did not show a significant difference between predicted GTV (CNN-Rad) and GTV (CNN-Histo) ground truth (p = 0.18).Discussion and Conclusion
In this study, mpMRI was combined with whole mount histopathology slices to evaluate the quality of a CNN for GTV segmentation. The segmentation quality of the CNN is comparable to that of similar studies with a DSC of 0.3515. CNNs are usually trained on the tumor segmentations drawn in accordance with the PI-RADS protocol 16 on the MRI. It is not clear, however, if the network learns to discriminate healthy from diseased tissue, or if it just learns to reproduce the experts work, including the systematic biases. Testing the segmentation performance against radiologist-based and histology-based ground truth, we could not find a significant difference in the segmentation. These results indicate that the network can discriminate tumor from healthy tissue even under a training with imperfect ground truth data. Increasing the data base with segmentations from multiple readers might further improve the CNN as systematic biases from a single reader could be averaged out. In general, the use of manually segmented ground truth data is justified for the training of CNNs for PCa segmentation. This is especially critical, as there cannot be any other ground truth for patients who do not undergo prostatectomy due to better, less invasive treatment options, e.g. if the tumor is curable by radiation therapy.Acknowledgements
Grant support by the Klaus Tschira Stiftung GmbH, Heidelberg, Germany is gratefully acknowledged.References
- H. U. Ahmed et al., “Diagnostic accuracy of multi-parametric MRI and TRUS biopsy in prostate cancer (PROMIS): a paired validating confi rmatory study,” Lancet, vol. 389, pp. 815–822, 2017.
- P. Steenbergen et al., “Prostate tumor delineation using multiparametric magnetic resonance imaging : Inter-observer variability and pathology validation,” Radiother. Oncol., vol. 115, no. 2, pp. 186–190, 2015.
- L. Rundo et al., “CNN-Based Prostate Zonal Segmentation on T2-Weighted MR Images: A Cross-Dataset Study,” Smart Innov. Syst. Technol., vol. 151, pp. 269–280, 2020.
- S. Motamed, I. Gujrathi, D. Deniffel, A. Oentoro, M. A. Haider, and F. Khalvati, “A Transfer Learning Approach for Automated Segmentation of Prostate Whole Gland and Transition Zone in Diffusion Weighted MRI,” 2019.
- S. Hu, D. Worrall, S. Knegt, B. Veeling, H. Huisman, and M. Welling, “Supervised Uncertainty Quantification for Segmentation with Multiple Annotations,” in Lecture Notes in Computer Science (including subseries Lecture Notes in Artificial Intelligence and Lecture Notes in Bioinformatics), 2019, vol. 11765 LNCS, pp. 137–145.
- D. Karimi, G. Samei, C. Kesch, G. Nir, and S. E. Salcudean, “Prostate segmentation in MRI using a convolutional neural network architecture and training strategy based on statistical shape models,” Int. J. Comput. Assist. Radiol. Surg., vol. 13, no. 8, pp. 1211–1219, Aug. 2018.
- G. Litjens et al., “Evaluation of prostate segmentation algorithms for MRI: The PROMISE12 challenge,” Med. Image Anal., vol. 18, no. 2, pp. 359–373, Feb. 2014.
- K. Gennaro, K. Porter, J. Gordetsky, S. Galgano, and S. Rais-Bahrami, “Imaging as a Personalized Biomarker for Prostate Cancer Risk Stratification,” Diagnostics, vol. 8, no. 4, p. 80, Nov. 2018.
- L. Boesen, E. Chabanova, V. Løgager, I. Balslev, and H. S. Thomsen, “Apparent diffusion coefficient ratio correlates significantly with prostate cancer gleason score at final pathology,” J. Magn. Reson. Imaging, vol. 42, no. 2, pp. 446–453, Aug. 2015.
- S. S. Salami, E. Ben-Levi, O. Yaskiv, B. Turkbey, R. Villani, and A. R. Rastinehad, “Risk stratification of prostate cancer utilizing apparent diffusion coefficient value and lesion volume on multiparametric MRI,” J. Magn. Reson. Imaging, vol. 45, no. 2, pp. 610–616, 2017.
- J. O. Barentsz et al., “Synopsis of the PI-RADS v2 Guidelines for Multiparametric Prostate Magnetic Resonance Imaging and Recommendations for Use,” European Urology, vol. 69, no. 1. Elsevier B.V., pp. 41–49, 01-Jan-2016.
- C. Zamboglou et al., “The impact of the co-registration technique and analysis methodology in comparison studies between advanced imaging modalities and whole-mount-histology reference in primary prostate cancer,” Sci. Rep., vol. 11, no. 1, pp. 1–11, 2021.
- O. Ronneberger, P. Fischer, and T. Brox, “U-net: Convolutional networks for biomedical image segmentation,” in International Conference on Medical Image Computing and Computer-Assisted Intervention, 2015, pp. 234–241.
- A. S. Bettermann et al., “[(68)Ga-]PSMA-11 PET/CT and multiparametric MRI for gross tumor volume delineation in a slice by slice analysis with whole mount histopathology as a reference standard - Implications for focal radiotherapy planning in primary prostate cancer.,” Radiother. Oncol. J. Eur. Soc. Ther. Radiol. Oncol., vol. 141, pp. 214–219, Dec. 2019.
- P. Schelb et al., “Classification of cancer at prostate MRI: Deep Learning versus Clinical PI-RADS Assessment,” Radiology, vol. 293, no. 3, pp. 607–617, 2019. [16] J. O. Barentsz et al., “ESUR prostate MR guidelines 2012,” Eur. Radiol., vol. 22, no. 4, pp. 746–757, 2012.
Figures
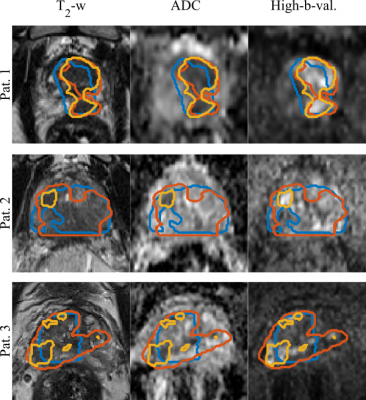
GTV Segmentations
overlaid on the input image sequences for 3 patients from the test set.Ground truth segmentations GTV-Histo (blue),
GTV-MRI (orange) and the predicted segmentation GTV-CNN (yellow).
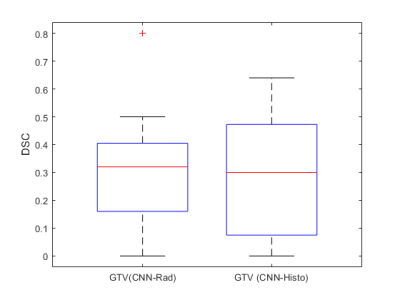
The
DSC for GTV (CNN-Rad) and GTV(CNN-Histo ) on the test cohort (n=15).The red
lines in the plot show the median DSC. The upper and lower bounds of the blue
box indicate the 25th and 75th percentiles, respectively.
DOI: https://doi.org/10.58530/2022/0925