0886
Brain tissue segmentation on 3D-FLAIR weighted images in multiple sclerosis1Anatomy and Neurosciences, MS Center Amsterdam, Amsterdam Neuroscience, Amsterdam UMC, Vrije Universiteit Amsterdam, Amsterdam, Netherlands, 2Radiology and Nuclear Medicine, MS Center Amsterdam, Amsterdam Neuroscience, Amsterdam UMC, Vrije Universiteit Amsterdam, Amsterdam, Netherlands, 3Neurology, MS Center Amsterdam, Amsterdam Neuroscience, Amsterdam UMC, Vrije Universiteit Amsterdam, Amsterdam, Netherlands, 4Neurology and Healthcare Engineering, UCL London, Amsterdam, United Kingdom, 5Biomedical Engineering & Physics, Amsterdam UMC, location AMC, Amsterdam, Netherlands
Synopsis
Conventional brain segmentation approaches typically require high resolution 3D-T1 weighted images, which are often unavailable in clinical multiple sclerosis (MS) protocols. Recently, SynthSeg was released which allows 3D-FLAIR to be used for segmentations. This study compared segmentation on 3D-FLAIR with SynthSeg to segmentation in the same patients on 3D-T1 (SynthSeg, FastSurfer and SAMSEG). Brain segmentation was performed on 100 patients with 3D-T1 and 3D-FLAIR images from research and clinical datasets. ICC results showed good comparability for brain tissue, ventricle and grey matter assessments in research data. In clinical data only good comparability was found for ventricle segmentation.
Introduction
Accelerated brain tissue loss occurs from the earliest stages in multiple sclerosis (MS) and is associated with disability and cognitive impairment.1 Brain volumetric measurements on MRI provide important imaging markers for disease progression and evaluating therapeutic effects.2 To date, these imaging markers are primarily used as outcome measures in research and clinical trials. Clinical protocols commonly focus on 3D-FLAIR over 3D-T1 imaging, while most segmentation methods are only compatible with the latter.3 To overcome this issue, new segmentation approaches have been developed that work on other MRI contrasts and lower resolutions. The recently developed convolutional neural network (CNN)-based segmentation software SynthSeg showed promising results in brain tissue segmentation of contrasts other than 3D-T1, but performance on 3D-FLAIR remains to be validated.4 The aim of this study was to investigate agreement between 3D-T1 weighted brain volumetrics and SynthSeg-based volumetrics on 3D-FLAIR .Methods
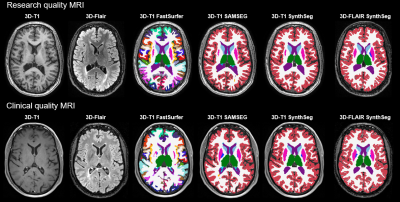
To evaluate the agreement of brain volumetric measurements on 3D-T1 and 3D-FLAIR images, 100 MRI examinations from 100 MS patients were selected. MRI data were acquired on two 1.5 T systems (Siemens Magnetom Sola, Siemens Avanto) and three 3.0 T systems (Siemens Magnetom Vida, GE Discovery MR750, GE Signa HDxT). From each scanner, 20 MRI examinations were included. The dataset contained both research and clinical datasets. Research imaging protocols contained a 3D-T1 weighted sequence (FSPGR) without contrast and 3D-T2 weighted FLAIR sequence. Clinical imaging protocols contained a 3D-T1 weighted sequence with or without contrast (T1 SPACE FS, T1 FLASH) and a 3D-T2 weighted FLAIR sequence. Segmentation was performed on 3D-T1 weighted images using FastSurfer, SAMSEG (cross-sectional pipeline for MS patients) and SynthSeg.4-6 3D-FLAIR weighted images were segmented with SynthSeg only. Brain, ventricular, cortical grey matter (GM), total deep grey matter (DGM) and thalamic volumes were compared between 3D-T1 and 3D-FLAIR based segmentations with calculation of the intraclass correlation coefficient (ICC; two way-mixed model, absolute agreement). Thalamic volume was assessed in addition to DGM given its known strong clinical predictive value in MS.7 Analyses were performed separately on the research data (n=40, GE scanners) and the clinical data (n=60, Siemens scanners). An example of a research and a clinical MRI exam with their corresponding segmentations are shown in Figure 1.Results
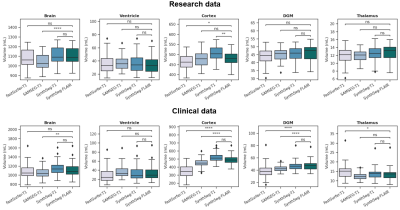
In the research data, good (ICC>0.8) agreement was found between 3D-FLAIR segmentation and all 3D-T1 segmentations for most assessed brain structures (Figure 2). Only poor agreement was found for SAMSEG on brain and thalamic volumes. In clinical data, ICC calculations showed good agreement between SynthSeg on 3D-FLAIR and SynthSeg on 3D-T1 for all measures, i.e. brain, ventricle, cortex, deep grey matter and thalamic volumes (Figure 2). However, agreement between SynthSeg on 3D-FLAIR and 3D-T1-based FastSurfer and SAMSEG segmentation was poor for cortical and deep grey matter volumes. FastSurfer and SAMSEG on 3D-T1 displayed lower volumes of these structures compared to the 3D-Flair segmentation (Figure 3). Total brain volume agreement for FastSurfer with SynthSeg on 3D-Flair was good, but poor for SAMSEG. Only ventricular volumes were highly comparable between methods in the clinical dataset. FastSurfer missed parts of the brain in some cases with poor grey/white matter contrast, an example is shown in Figure 4.Discussion
In this study, SynthSeg was used to determine volumetric measurements on 3D-FLAIR images, which were compared to 3D-T1 measurements using three automated segmentation approaches. SynthSeg achieved high agreement between 3D-T1 and 3D-FLAIR based segmentation in both research and clinical datasets. However, specific variations in agreement were observed, showing especially a lower agreement with SAMSEG. The segmentations of FastSurfer and SAMSEG may not have been the best reference in the clinical dataset, since these methods typically require high quality images with sufficient grey matter/white matter contrast, which might not always be the case for the 3D-T1 in clinical protocols. In the 3D-T1 and 3D-FLAIR images from research protocols, a high agreement was found between 3D-T1 segmentations (FastSurfer and SynthSeg) and SynthSeg 3D-FLAIR segmentation for all structures, while SAMSEG showed poorer agreement. Therefore, 3D-FLAIR segmentation with SynthSeg is a promising approach for volumetric measurements in MS patients, but further validation is required.Conclusion
Brain, cortical, ventricular and deep grey matter volume assessments on 3D-FLAIR weighted images with SynthSeg show good agreement to 3D-T1 segmentation with FastSurfer and SynthSeg in high quality research data. In clinical data only ventricular volume could be reliably measured on 3D-FLAIR weighted images in MS patients. Future studies should validate this finding to enable FLAIR-based volumetric measurements in clinically acquired MRI data.Acknowledgements
No acknowledgement found.References
1. Roosendaal SD, Bendfeldt K, Vrenken H, et al. Grey matter volume in a large cohort of MS patients: relation to MRI parameters and disability. Multiple Sclerosis Journal 2011;17:1098-1106.
2. De Stefano N, Airas L, Grigoriadis N, et al. Clinical relevance of brain volume measures in multiple sclerosis. CNS drugs 2014;28:147-156.
3. Rocca MA, Battaglini M, Benedict RH, et al. Brain MRI atrophy quantification in MS: from methods to clinical application. Neurology 2017;88:403-413.
4. Billot B, Greve DN, Puonti O, et al. SynthSeg: Domain Randomisation for Segmentation of Brain MRI Scans of any Contrast and Resolution. ArXiv 2021;abs/2107.09559.
5. Cerri S, Puonti O, Meier DS, et al. A contrast-adaptive method for simultaneous whole-brain and lesion segmentation in multiple sclerosis. NeuroImage 2021;225:117471.
6. Henschel L, Conjeti S, Estrada S, Diers K, Fischl B, Reuter M. Fastsurfer-a fast and accurate deep learning based neuroimaging pipeline. NeuroImage 2020;219:117012.
7. Eshaghi A, Prados F, Brownlee WJ, et al. Deep gray matter volume loss drives disability worsening in multiple sclerosis. Annals of neurology 2018;83:210-222.
Figures