0852
Brain Tumor Segmentation Using 3D CMM-Net with Limited and Accessible MR Images1Department of Electrical and Electronic Engineering, Yonsei University, Seoul, Korea, Republic of
Synopsis
In this work, we investigate the possibility of using a limited number of medical MR images for brain tumor segmentation. This can assist to perform the segmentation task using limited and accessible data in clinical practice. We observed that the FLAIR and T1CE are the most suitable images that maintain comparable segmentation performances similar to the case of using more data (i.e., T1, T2, FLAIR, and T1CE). To further improve the segmentation results, we augment the selected image pair and generate an additional fusion map using the Singular Value Decomposition (SVD).
Introduction
Magnetic Resonance Imaging (MRI) plays an important role in brain tumors diagnosis. This is due to the possibility of MRI to generate several soft-tissue images, such as T1, T2, FLAIR, and T1CE. However, it is difficult to obtain all types of MR images in actual clinical situations due to the time and cost required to acquire a complete set. In this work, we investigate the possibility of using a limited number of medical MR images for brain tumor segmentation. This can assist to perform the segmentation task using limited and accessible data in clinical practice. Further, we augment the selected image pair and generate an additional fusion map using the SVD. This generated fusion map may contain valuable information and patterns, and hence, assist to improve the segmentation of brain tumors.Methods
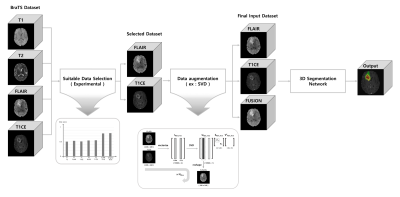
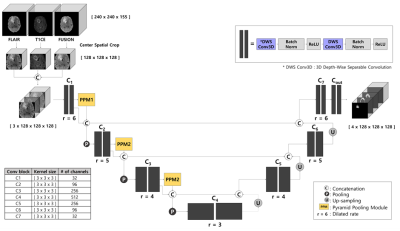
An overview diagram of the proposed work is illustrated in Figure 1.
[Network]
We employed the Contextual Multi-Scale Multi-Level Network (CMM-Net)1 as our backbone since it has an attractive advantage of segmentation tasks in various medical domains. In this work, we developed a 3D version of the existing 2D CMM-Net. Instead of using the conventional 3D convolution, we exploited the usage of Depth-Wise Separable Convolution (DWS Conv)2, which not only can reduce the number of parameters by 20 times in our case but also maintain the performance compared to the conventional one. We also applied dilated convolution to the whole convolution blocks in the proposed network. This allows the network to learn a larger receptive field without increasing the number of training weights3. Also, the Pyramid Pooling Module (PPM) was utilized in the encoder part to derive multi-scale feature maps4. The network architecture is presented in Figure 2.
[Selection of Most Suitable Images and Augmentation]
The procedure of selecting the most suitable MR images for the task of brain tumor segmentation is achieved experimentally by feeding different image pairs, that contain various combinations of BraTS images, into our proposed 3D CMM-Net. More specifically, we investigate which image combination (T1&T2, T1&FLAIR, T1&T1CE, T2&FLAIR, T2&T1CE, or FLAIR&T1CE) contains significant information through monitoring the overall segmentation result compared to the usage of four images of the BraTS dataset. This procedure enables to accomplish the segmentation task using a limited number of MR images and becomes the more practical solution for the majority of clinical facilities that acquire limited types of MR sequences.
For further improvement of the results that were obtained using only the selected pair image, we perform data augmentation. In general, there are many augmentation methods in literature5. In this work, we generate new samples as a linear combination of the selected images with larger singular value using the Singular Value Decomposition (SVD) method. We called this new augmented image as fusion map.
We cropped all datasets using the center spatial crop from 240×240×155 to 128×128×128 voxels after normalization. This cropping process enables to reduce the size of input images and hence maintains lower computation costs during training. To take the advantage of the presence of three different image modalities (i.e., FLAIR, T1CE, and FUSION), we concatenated all three images and utilized them as an input (Fusion_3ch) to our network as shown in Figure 2.
Results
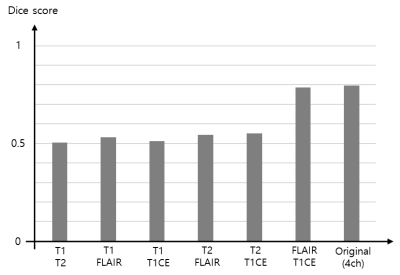
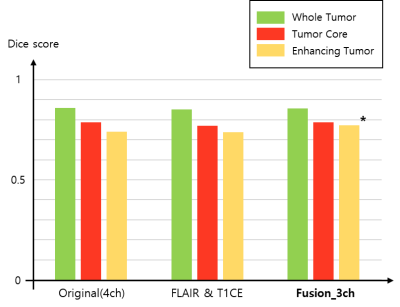
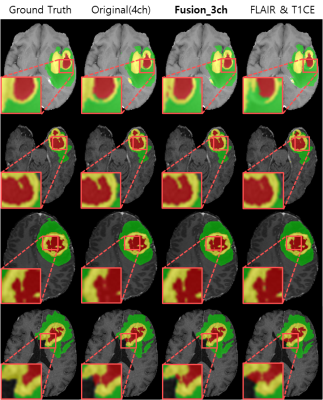
For the experiments, we used the BraTS2021 training dataset composed of 1,251 patients. We randomly selected 51 patients for the test and the remaining 1,200 patients were utilized for network training6-8. To find out which image pair has the most useful information for the brain tumor segmentation (i.e., using only two channels as an input), we trained the model using six different combinations as shown in Figure 3. The segmentation results of using the image pairs of FLAIR and T1CE outperformed the other combinations and achieved an average Dice score similar to the results of using the original four inputs. Figure 4 presents the quantitative results for segmentation performance using the different data combinations: Original of BraTS (4ch), FLAIR and T1CE, and Fusion_3ch. Overall, the segmentation performance in the case of using the Fusion_3ch gets improved compared to the using of only the FLAIR and T1CE. We observe that the enhancing tumor class has the largest improvement rate of 1.7% compared to other tumors.We also presented the qualitative result in Figure 5. The segmentation result from the Fusion_3ch dataset is similar to the ground-truth compared to the others, especially for the enhancing tumor regions.
Discussion and Conclusion
In this work, we examined the most suitable image pair for the task of brain tumor segmentation when using 2ch images as an input. We found that it is advantageous for our task to use images that adequately present the characteristics of the tumor than utilizing images with a large amount of redundant information. Also, we proposed an approach for generating the composite fusion image from the selected FLAIR and T1CE using the SVD.As a result, we obtained an enhancement in the performance of brain tumor segmentation, especially for the enhancing tumor region through the incorporating of the generated fusion map as an input compared to the using of the original BraTS dataset.
Acknowledgements
This work was supported by the Korea Medical Device Development Fund grant funded by the Korea government (the Ministry of Science and ICT, the Ministry of Trade, Industry and Energy, the Ministry of Health & Welfare, Republic of Korea, the Ministry of Food and Drug Safety) (Project Number: 202011D23)References
1. Al-Masni MA, Kim D-H. CMM-Net: Contextual multi-scale multi-level network for efficient biomedical image segmentation. Scientific reports 2021;11(1):1-18.
2. Chollet F. Xception: Deep learning with depthwise separable convolutions. 2017. p 1251-1258.
3. Yu F, Koltun V. Multi-scale context aggregation by dilated convolutions. arXiv preprint arXiv:151107122 2015.
4. Zhao H, Shi J, Qi X, Wang X, Jia J. Pyramid scene parsing network. 2017. p 2881-2890.
5. Perez L, Wang J. The effectiveness of data augmentation in image classification using deep learning. arXiv preprint arXiv:171204621 2017.
6. Baid U, Ghodasara S, Mohan S, Bilello M, Calabrese E, Colak E, Farahani K, Kalpathy-Cramer J, Kitamura FC, Pati S. The rsna-asnr-miccai brats 2021 benchmark on brain tumor segmentation and radiogenomic classification. arXiv preprint arXiv:210702314 2021.
7. Lloyd CT, Sorichetta A, Tatem AJ. High resolution global gridded data for use in population studies. Scientific data 2017;4(1):1-17.
8. Menze BH, Jakab A, Bauer S, Kalpathy-Cramer J, Farahani K, Kirby J, Burren Y, Porz N, Slotboom J, Wiest R. The multimodal brain tumor image segmentation benchmark (BRATS). IEEE transactions on medical imaging 2014;34(10):1993-2024.
Figures