0850
Distortion Free Image Reconstruction using a Deep Neural Network for an MRI-Linac1ACRF Image X Institute, University of Sydney, Sydney, Australia, 2Ingham Institute For Applied Medical Research, Sydney, Australia, 3School of Information Technology and Electrical Engineering, University of Queensland, Brisbane, Australia
Synopsis
The advent of MRI-guided radiotherapy has elevated demand for high geometric fidelity imaging. However, gradient nonlinearity can cause image distortion, which limits the accuracy of radiotherapy. In this work, we develop a deep neural network, namely DFReconNet, to reconstruct distortion free images directly from raw k-space in real time. Experiments on simulated brain datasets and phantom images acquired from an MRI-Linac demonstrated the utility of the proposed method.
Introduction
In conventional clinical MRI, images are reconstructed under the assumption that linear spatial gradient encoding has operated on MR signals [1]. However, the engineering constraints on gradient coil performance and system efficiency make generating linear gradient fields across the entire field of view (FOV) impractical [2]. The presence of gradient field nonlinearity (GNL) causes image distortions and is particularly problematic for MRI-guided radiotherapy treatment where anatomy must be localized with sub millimetre precision [3]. Image distortions can be corrected by image-domain interpolation [1] or k-space domain reconstruction [2] method. However, computational complexity limits their application for real-time tumor tracking during treatments [4]. Our recently developed ReUINet method shows the promise of deep neural networks for fast distortion correction in the image domain [5]. In this work, to further reduce the computational cost, we propose a novel deep learning network, DFReconNet, that can reconstruct distortion free (DF) images directly from the k-space data. Simulated brain dataset and experimental phantom images acquired from an MRI-Linac were used to investigate the proposed method.Methods
Problem formulationThe forward encoding process with gradient nonlinearity can be formulated as [6]: $$ E_{GNL}\cdot Fm=b\;\;\;\;\;\;\;\;\;\;\;\;\;\;\;\;\;\;\;\;\;\;\;\;\;\;\;\;\;\;\;\;\;\;(1)$$
Where b is the GNL-corrupted k-space signal; F represents theoretical Fourier transform matrix and m denotes the distortion free image. EGNL is the GNL encoding matrix with the kernel of eGNL =e-2πjkΔ(L), where Δ(L) is the GNL induced spatial deviation at location L. The distortion free image can be reconstructed by the penalized regression [2]:$$m=\mathop{\arg\min}_{\ m} \{\lambda P(m)+\| E_{GNL} \cdot Fm-b\|_F^2\} \;\;\;\;\;\;\;\;\;\;\;\;\;\;\;\;\;(2)$$ where P(m) denotes a regularization function with a weighting parameter λ. Optimization algorithms, e.g., nonlinear conjugate gradients, can be used to solve Eq. (2); however, they normally come with the compromise of high computational time.
DFReconNet
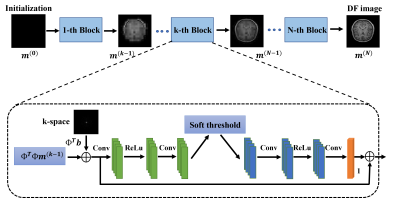
Inspired by an interpretable optimization-based network, i.e., ISTA-Net [7], we propose a distortion free reconstruction network (DFReconNet) to solve the minimization problem in Eq. (2). As shown in Figure. 1, DFReconNet consists of four iterative blocks and each block learns one iteration in ISTA algorithm. Each block starts with a data fidelity calculation, followed by a learnable nonlinear forward transform, a soft-thresholding operation, and a learnable nonlinear backward transform. The forward and backward transforms in each block were learned using the combination of a rectified linear unit (ReLU) and two convolutional layers.
Data preparation and network training
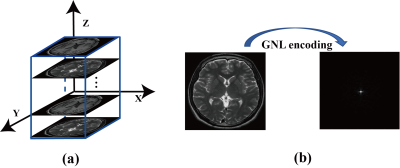
In this study, 3000 T1-weighted brain images from a public MR dataset [8] were selected as training labels. Brain images were acquired with a whole-body MRI scanner and the imaging parameters were: voxel size = 320 × 320 × 256, resolution = 0.7 mm × 0.7 mm × 0.7 mm and TE/TR = 2.13 ms/2.4 s. Twenty locations with uniform intervals (15 mm) were allocated along z direction in a FOV of 30 cm × 30 cm × 30 cm, as shown in Figure 2. Every 150 randomly selected brain images were positioned at each location and our previously developed GNLNet [4] was used to provide the GNL field information. The corresponding GNL-corrupted k-space data of each brain image was generated by using Eq. (1). Repeated operations were conducted for the other two orthogonal planes. The proposed DFReconNet was trained on an Nvidia Tesla V100 GPU (32G) for 100 epochs (~10 hours) using these simulated images with Adam optimizer.
Another 300 brain images from the same dataset were used to prepare the simulated testing data. A 3D grid phantom was scanned from the 1.0T Australian MRI-Linac system with the imaging paramters: matrix size = 130 × 110 × 192, resolution = 1.8 mm × 2 mm × 1.8 mm, and TE/TR = 15 ms/5.1 s. 192 phantom slices were obtained to validate the proposed method.
Results
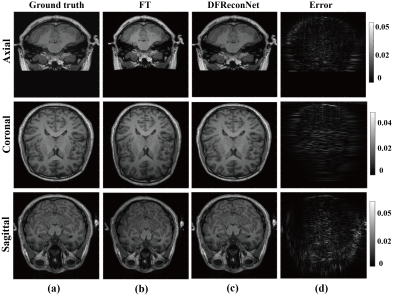
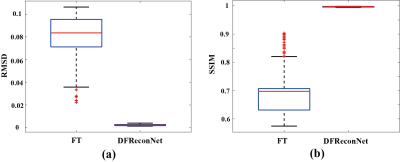
Figure 3 shows brain results reconstructed by standard Fourier transform (FT) and the proposed DFReconNet at three orthogonal planes. Compared with the ground truth images, considerable geometric distortions including image shrinkage (axial and sagittal) and dilation (coronal) are presented in Figure 3(b). By contrast, the DFReconNet successfully eliminated these distortions and resulted in negligible errors (less than 5%). The root mean square deviation (RMSD) and structural similarity index (SSIM) values were calculated on 300 testing brain images. As shown in Figure 4, the median RMSD of FT-reconstructed images is ~0.08, which is approximately 8-fold greater than that of DFReconNet results (~0.01). Similarly, increased SSIM median value was observed when using DFReconNet (~0.98) in comparison to FT method (~0.7).Figure 5 illustrates the DFReconNet results on phantom images. Compared to the FT method, geometric distortions are successfully reduced in DFReconNet-reconstructed images, which is consistent with the results of Figure 3. The reconstruction time of the proposed network on an image of size 128 × 128 is 0.08s with an Nvidia Tesla V100 GPU (32G), which makes the proposed method feasible for real-time imaging applications.
Discussion and conclusion
Results on simulated brain dataset and experimental phantom images indicated that the proposed method was capable of reconstructing distortion free images directly from k-space domain in real-time, which can facilitate the accurate radiotherapy treatment on an MRI-Linac.Acknowledgements
The authors acknowledge the financial support of the NHMRC grant (grant No. 1132471) - The Australian MRI-Linac Program: Transforming the Science and Clinical Practice of Cancer Radiotherapy.
David Waddington and Paul Liu are supported by the Cancer Institute NSW. Brendan Whelan is supported by NHMRC CJ Martin fellowship.
References
[1]. Tao S, Trzasko JD, Gunter JL, et al. Gradient nonlinearity calibration and correction for a compact, asymmetric magnetic resonance imaging gradient system. Phys Med Biol. 2017;62(2):N18-N31.
[2]. Tao S, Trzasko JD, Shu Y, Huston III J, Bernstein MA. Integrated image reconstruction and gradient nonlinearity correction. Magnetic resonance in medicine. 2015;74(4):1019-1031.
[3]. Shan, S., Liney, G. P., Tang, F., Li, M., Wang, Y., Ma, H., ... & Crozier, S. (2020). Geometric distortion characterization and correction for the 1.0 T Australian MRI‐linac system using an inverse electromagnetic method. Medical physics, 47(3), 1126-1138.
[4]. Li, M., Shan, S., Chandra, S.S., Liu, F. and Crozier, S., 2020. Fast geometric distortion correction using a deep neural network: Implementation for the 1 Tesla MRI‐Linac system. Medical Physics, 47(9), pp.4303-4315.
[5]. Shan, S., Li, M., Li, M., Tang, F., Crozier, S. and Liu, F., 2021. ReUINet: A fast GNL distortion correction approach on a 1.0 T MRI‐Linac scanner. Medical Physics.
[6]. Shan S, Li M, Tang F, Ma H, Liu F, Crozier S. Gradient Field Deviation (GFD) Correction Using a Hybrid-Norm Approach With Wavelet Sub-Band Dependent Regularization: Implementation for Radial MRI at 9.4 T. IEEE Transactions on Biomedical Engineering. 2019;66(9):2693-2701.
[7]. Zhang, J. and Ghanem, B., 2018. ISTA-Net: Interpretable optimization-inspired deep network for image compressive sensing. In Proceedings of the IEEE conference on computer vision and pattern recognition (pp. 1828-1837). 8. Sunavsky, A. and Poppenk, J., 2020. Neuroimaging predictors of creativity in healthy adults. Neuroimage, 206, p.116292.
Figures