0834
Assessment of Lipid Biosynthesis and Turnover in Renal Cell Carcinoma Cells and Tissues Using NMR-Resolved Stable Isotope Experiments
Daniel Robert Crooks1, Ye Yang2, Andrew Lane3, Teresa Fan3, Jeffrey Brender4, Murali C Krishna4, and W. Marston Linehan1
1Urologic Oncology Branch, National Cancer Institute, Bethesda, MD, United States, 2National Cancer Institute, Bethesda, MD, United States, 3Center for Environmental Systems Biochemistry, University of Kentucky, Lexington, KY, United States, 4Radiation Biology Branch, National Cancer Institute, Bethesda, MD, United States
1Urologic Oncology Branch, National Cancer Institute, Bethesda, MD, United States, 2National Cancer Institute, Bethesda, MD, United States, 3Center for Environmental Systems Biochemistry, University of Kentucky, Lexington, KY, United States, 4Radiation Biology Branch, National Cancer Institute, Bethesda, MD, United States
Synopsis
NMR-based analyses of lipids can shed insight into the global complement of cellular lipids, and elucidate the fuel sources and pathways contributing to lipid biosynthesis in cells grown in the presence of 13C-labeled fuels. We utilized 1H-13C HSQC NMR analysis of cellular lipids derived from FH- and mtDNA deficient UOK271 cells and observed robust reductive carboxylation of glutamine that resulted in formation and incorporation of 13C-glutamine-derived acetyl groups into lipid chains.
Summary:
We utilized 1H-13C HSQC NMR to analyze the fuel sources for lipid biosynthesis in patient-derived fumarate hydratase (FH) and mtDNA deficient UOK271 cells and found that 13C-glucose labeled lipid glycerol moieties but not acyl chains, while acyl chains were robustly labeled by 13C-glutamine.
Introduction:
Hereditary leiomyomatosis and renal cell carcinoma (HLRCC)-associated fumarate hydratase (FH)-deficient renal cell carcinoma (RCC) is the most aggressive and lethal of the 14 known types of genetically defined heritable forms of RCC 1-3. Previous work has demonstrated glucose-dependent growth as well as up-regulation of the pentose phosphate pathway (PPP) providing cytosolic reducing equivalents for anabolic reactions including lipid biosynthesis 2,4. Recently, we identified loss and mutation of mitochondrial DNA as a hallmark of human FH-deficient tumors 5, demonstrating that this tumor type undergoes a permanent departure from mitochondrial oxidative phosphorylation. FH-deficient tumor cells are able to multiply and metastasize with striking rapidity, a process which involves a massive amount of lipid and membrane biosynthesis. The enormous chemical diversity of cellular lipids makes their detailed analysis very complex. NMR analysis of lipids offers a unique modality to assess the global isotope labeling patterns of cellular lipids in cells and tissues grown in the presence of stable isotope tracers 6,7. In this study, we have utilized stable isotope-resolved metabolomics and NMR spectroscopy to evaluate sources of de novo lipid biosynthesis in FH- and mtDNA-deficient UOK271 tumor cells.
Methods:
Tracer experiments were carried out in triplicate with either 10 mM 13C6-glucose or 2mM 13C515N2-L-glutamine in DMEM containing 10% dialyzed FBS and 1mM sodium pyruvate for 48 hours. At the end of the tracing period, cells were extracted in acetonitrile:water:chloroform (2:1.5:1) 8.A 1:1 mixture of chloroform and methanol containing 1 mM butylated hydroxytoluene (BHT) was added to the lower non-polar fractions obtained from the extracts described above, and the mixtures were then dried in an Eppendorf SpeedVac. The resulting lipid residues were dissolved in 210 µL of d4-methanol, vortexed vigorously, centrifuged at 20,000 x g for 5 minutes, and 200 µL of the supernatant was transferred to disposable 3 mm Bruker SampleJet glass NMR tubes. NMR spectra were recorded at 700Mhz with a triple inverse resonance cold probe. 1H{13C}-HSQC spectra were recorded with an acquisition time of 0.2 s and a relaxation delay of 1.8 s, with adiabatic decoupling. 1H-13C HSQC NMR spectra were referenced to the natural abundance 1H-13C BHT tert-butyl resonance at 1.40 ppm. 1D HSQC experiments were used to estimate the relative 13C content of different lipid functional group positions in the extracts, and peak areas were quantified where indicated and normalized to the methanol resonance.
Results:
Incubation of UOK271 cells in the presence of 10 mM 13C6-glucose resulted in robust labeling of the glycerol resonances in lipids at 5.24, 4.45, 4.43, 4.17, and 4.00 ppm as measured by 1H{13C}-HSQC, indicating robust incorporation of glucose-derived carbon into the glycerol moiety of cellular lipids (Figure 1). However, resonances corresponding to acyl chain carbons and terminal omega methyl groups in 13C6-glucose-labeled cells were comparable in intensity to spectra obtained from unlabeled cells (Figure 1). In sharp contrast, UOK271 cells grown in the presence of 2mM 13C515N2-L-glutamine did not show appreciable labeling of the glycerol moiety of cellular lipids, but did show significant labeling of lipid acyl chain resonances (1.29ppm), including those proximal to the lipid head group (2.33 and 1.60ppm), as well as terminal ω-methyl groups (0.9ppm). Analysis of the fatty acid synthesis precursor molecule citrate by mass spectrometry demonstrated that isotopic labeling of citrate by 13C6-glucose was minimal in UOK271 cells, whereas growth of the cells in the presence of 13C515N2-L-glutamine resulted in robust labeling of citrate with five additional mass units (m+5; Figure 2). Notably, restoration of fumarate hydratase enzyme activity in UOK271 cells by stable transfection of wild type human FH enzyme did not result in a significant change in 13C515N2-L-glutamine labeling of acyl chains, where empty vector-transfected UOK271 cells showed a acyl chain peak area of 431±91 while FH-transfected UOK271 cells showed a peak area of 408±87 (methanol-normalized; P=0.65, 2-tailed t-test).
Discussion:
Our data demonstrate that FH- and mtDNA-deficient UOK271 tumor cells utilized glutamine-derived carbon to de novo synthesize new cellular lipid chains, while glucose-derived carbon was used to synthesize the glycerol backbone needed for lipids. A profound lack of lipid acyl chain labeling from 13C6-glucose suggests that glucose did not appreciably enter the Krebs cycle, and concordantly citrate labeling was absent in UOK271 cells grown in the presence of 13C6-glucose. In contrast, growth of UOK271 cells in the presence of 13C515N2-L-glutamine resulted in robust 13C labeling of lipid acyl chains, demonstrating that this nutrient is important for lipid metabolism in these cells. The prominent m+5 isotopologue labeling pattern of citrate grown in the presence of 13C515N2-L-glutamine strongly suggests that citrate was derived by reductive carboxylation of glutamine-derived α-ketoglutarate, a process that has been observed in other models of FH-deficient RCC 9. Notably, reductive carboxylation of glutamine-derived α-ketoglutarate leading to lipid biosynthesis persisted following restoration of FH enzyme activity in the cells, suggesting that their mtDNA deficiency strongly underlies the metabolic reprogramming observed in these tumor cells.
We utilized 1H-13C HSQC NMR to analyze the fuel sources for lipid biosynthesis in patient-derived fumarate hydratase (FH) and mtDNA deficient UOK271 cells and found that 13C-glucose labeled lipid glycerol moieties but not acyl chains, while acyl chains were robustly labeled by 13C-glutamine.
Introduction:
Hereditary leiomyomatosis and renal cell carcinoma (HLRCC)-associated fumarate hydratase (FH)-deficient renal cell carcinoma (RCC) is the most aggressive and lethal of the 14 known types of genetically defined heritable forms of RCC 1-3. Previous work has demonstrated glucose-dependent growth as well as up-regulation of the pentose phosphate pathway (PPP) providing cytosolic reducing equivalents for anabolic reactions including lipid biosynthesis 2,4. Recently, we identified loss and mutation of mitochondrial DNA as a hallmark of human FH-deficient tumors 5, demonstrating that this tumor type undergoes a permanent departure from mitochondrial oxidative phosphorylation. FH-deficient tumor cells are able to multiply and metastasize with striking rapidity, a process which involves a massive amount of lipid and membrane biosynthesis. The enormous chemical diversity of cellular lipids makes their detailed analysis very complex. NMR analysis of lipids offers a unique modality to assess the global isotope labeling patterns of cellular lipids in cells and tissues grown in the presence of stable isotope tracers 6,7. In this study, we have utilized stable isotope-resolved metabolomics and NMR spectroscopy to evaluate sources of de novo lipid biosynthesis in FH- and mtDNA-deficient UOK271 tumor cells.
Methods:
Tracer experiments were carried out in triplicate with either 10 mM 13C6-glucose or 2mM 13C515N2-L-glutamine in DMEM containing 10% dialyzed FBS and 1mM sodium pyruvate for 48 hours. At the end of the tracing period, cells were extracted in acetonitrile:water:chloroform (2:1.5:1) 8.A 1:1 mixture of chloroform and methanol containing 1 mM butylated hydroxytoluene (BHT) was added to the lower non-polar fractions obtained from the extracts described above, and the mixtures were then dried in an Eppendorf SpeedVac. The resulting lipid residues were dissolved in 210 µL of d4-methanol, vortexed vigorously, centrifuged at 20,000 x g for 5 minutes, and 200 µL of the supernatant was transferred to disposable 3 mm Bruker SampleJet glass NMR tubes. NMR spectra were recorded at 700Mhz with a triple inverse resonance cold probe. 1H{13C}-HSQC spectra were recorded with an acquisition time of 0.2 s and a relaxation delay of 1.8 s, with adiabatic decoupling. 1H-13C HSQC NMR spectra were referenced to the natural abundance 1H-13C BHT tert-butyl resonance at 1.40 ppm. 1D HSQC experiments were used to estimate the relative 13C content of different lipid functional group positions in the extracts, and peak areas were quantified where indicated and normalized to the methanol resonance.
Results:
Incubation of UOK271 cells in the presence of 10 mM 13C6-glucose resulted in robust labeling of the glycerol resonances in lipids at 5.24, 4.45, 4.43, 4.17, and 4.00 ppm as measured by 1H{13C}-HSQC, indicating robust incorporation of glucose-derived carbon into the glycerol moiety of cellular lipids (Figure 1). However, resonances corresponding to acyl chain carbons and terminal omega methyl groups in 13C6-glucose-labeled cells were comparable in intensity to spectra obtained from unlabeled cells (Figure 1). In sharp contrast, UOK271 cells grown in the presence of 2mM 13C515N2-L-glutamine did not show appreciable labeling of the glycerol moiety of cellular lipids, but did show significant labeling of lipid acyl chain resonances (1.29ppm), including those proximal to the lipid head group (2.33 and 1.60ppm), as well as terminal ω-methyl groups (0.9ppm). Analysis of the fatty acid synthesis precursor molecule citrate by mass spectrometry demonstrated that isotopic labeling of citrate by 13C6-glucose was minimal in UOK271 cells, whereas growth of the cells in the presence of 13C515N2-L-glutamine resulted in robust labeling of citrate with five additional mass units (m+5; Figure 2). Notably, restoration of fumarate hydratase enzyme activity in UOK271 cells by stable transfection of wild type human FH enzyme did not result in a significant change in 13C515N2-L-glutamine labeling of acyl chains, where empty vector-transfected UOK271 cells showed a acyl chain peak area of 431±91 while FH-transfected UOK271 cells showed a peak area of 408±87 (methanol-normalized; P=0.65, 2-tailed t-test).
Discussion:
Our data demonstrate that FH- and mtDNA-deficient UOK271 tumor cells utilized glutamine-derived carbon to de novo synthesize new cellular lipid chains, while glucose-derived carbon was used to synthesize the glycerol backbone needed for lipids. A profound lack of lipid acyl chain labeling from 13C6-glucose suggests that glucose did not appreciably enter the Krebs cycle, and concordantly citrate labeling was absent in UOK271 cells grown in the presence of 13C6-glucose. In contrast, growth of UOK271 cells in the presence of 13C515N2-L-glutamine resulted in robust 13C labeling of lipid acyl chains, demonstrating that this nutrient is important for lipid metabolism in these cells. The prominent m+5 isotopologue labeling pattern of citrate grown in the presence of 13C515N2-L-glutamine strongly suggests that citrate was derived by reductive carboxylation of glutamine-derived α-ketoglutarate, a process that has been observed in other models of FH-deficient RCC 9. Notably, reductive carboxylation of glutamine-derived α-ketoglutarate leading to lipid biosynthesis persisted following restoration of FH enzyme activity in the cells, suggesting that their mtDNA deficiency strongly underlies the metabolic reprogramming observed in these tumor cells.
Acknowledgements
Funding was supported by the intramural research program of the Center for Cancer Research, National Cancer Institute.References
1 Grubb, R. L. et al. Hereditary leiomyomatosis and renal cell cancer: a syndrome associated with an aggressive form of inherited renal cancer. J Urol 177, 2074-2080 (2007). 2 Linehan, W. M. et al. The Metabolic Basis of Kidney Cancer. Cancer Discov, doi:10.1158/2159-8290.CD-18-1354 (2019). 3 Tomlinson, I. P. et al. Germline mutations in FH predispose to dominantly inherited uterine fibroids, skin leiomyomata and papillary renal cell cancer. Nat Genet 30, 406-410 (2002). 4 Yang, Y. et al. Metabolic reprogramming for producing energy and reducing power in fumarate hydratase null cells from hereditary leiomyomatosis renal cell carcinoma. PLoS One 8, e72179, doi:10.1371/journal.pone.0072179 (2013). 5 Crooks, D. R. et al. Mitochondrial DNA alterations underlie an irreversible shift to aerobic glycolysis in fumarate hydratase-deficient renal cancer. Sci Signal 14, doi:10.1126/scisignal.abc4436 (2021). 6 Crooks, D. R. et al. Acute loss of iron-sulfur clusters results in metabolic reprogramming and generation of lipid droplets in mammalian cells. J Biol Chem 293, 8297-8311, doi:10.1074/jbc.RA118.001885 (2018). 7 Lin, P. et al. NMR Methods for Determining Lipid Turnover via Stable Isotope Resolved Metabolomics. Metabolites 11, doi:10.3390/metabo11040202 (2021).Figures
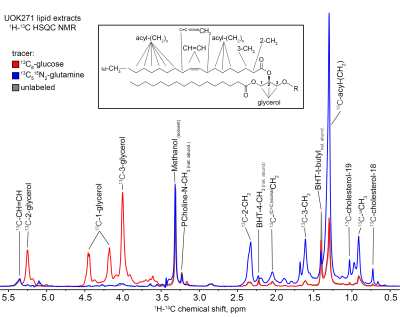
Figure 1: 1H-13C
HSQC NMR peaks corresponding to the terminal ω-methyl resonance (0.90 ppm), the
large saturated acyl methylene (e.g. C5-7,12-16) resonance (1.29 ppm), the 2-
and 3-methylene resonances of acyl chains (2.33, 1.60 ppm), the glycerol
resonances (5.24, 4.45, 4.43, 4.17, and 4.00 ppm), cholesterol methyl groups
(0.73 and 1.03 ppm), the unsaturated C=C double bond (5.35 ppm) and the
adjacent vicinal protons (2.04 ppm), and the natural abundance choline methyl
groups (3.23 ppm), solvent methanol 1H impurity resonance is observed
at 3.31 ppm.
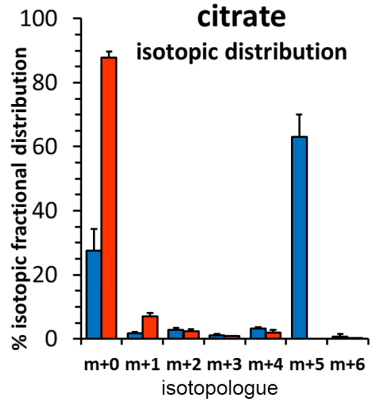
Figure 2: Citrate
isotopologues obtained from polar extracts of UOK271 cells labeled with either 13C6-glucose
(red bars) or 13C515N2-L-glutamine
(blue bars) were measured by GCMS. Data are expressed as % isotopic fractional distribution,
after correction for the isotope natural abundance.
DOI: https://doi.org/10.58530/2022/0834