0823
Enhanced T2-weighted images using Luminal Water Imaging and U-Net based segmentation for prostate cancer diagnosis.1UBC MRI Research Center, University of British Columbia, Vancouver, BC, Canada, 2Department of Physics and Astronomy, University of British Columbia, Vancouver, BC, Canada, 3Department of Urologic Sciences, University of British Columbia, Vancouver, BC, Canada, 4Vancouver Prostate Centre, Vancouver, BC, Canada, 5Department of Radiology, The University of British Columbia, Vancouver, BC, Canada
Synopsis
Clinical prostate carcinoma (PCa) detection with MRI has focused on qualitative assessment. However, novel Luminal Water Imaging (LWI) provides quantitative information with promising results for detecting PCa. To overcome shortcomings of clinical prostate MRI protocol, we propose using LWI to augment T2-weighted (T2W) images to improve image contrast for PCa detection while maintaining anatomical details needed for radiological diagnosis. Here, we investigate automatic segmentation models and various weighting functions of LWI parameter maps to generate semi-quantitative T2W images that also preserves anatomical detail. Our results show that a combined T2W and LWI parameter image provides enhanced detection of PCa.
Purpose
To investigate the automated enhancement of prostate cancer (PCa) detection in T2-weighted (T2W) images using Luminal Water Imaging (LWI) weighting and machine-learned segmentation.Introduction
Luminal Water Imaging is a novel, quantitative MRI technique developed for PCa detection. The technique measures the fractional volume of luminal water and therefore should provide signal differences between cancerous and non-cancerous prostate tissue1-3. LWI has already shown an equal or better correlation to Gleason Scores (GS) when compared with multi-parametric MRI (mp-MRI)2. However, these images (except T2W images) lack anatomical details that radiologist rely on for diagnosis. In this study, we investigate various weightings functions to best augment T2W images with LWI parametric maps to preserve anatomical information while improving PCa image contrast. Weighting is performed on regions-of-interest (ROIs) in T2W images, through automatic segmentation of the entire prostate and peripheral zone (PZ).Methods
MRI DataLWI data (240x240 pixels) and high-spatial-resolution T2W (512x512 pixels) images were collected for 40 patients with biopsy-proven PCa on a 3T MRI scanner (Philips Medical Systems) with protocols followed from a previous study2. From the LWI protocol, the multi-echo T2 signal decay curves were fitted using NNLS as implemented in DECAES4 and seven LWI parameter maps, each of size 240x240 pixels, were calculated1-3. The location and extent of tumours on the parametric maps were mapped from whole-mount histology using registration software developed in-house5. ROIs of the entire prostate and PZ in the multi-echo images were outlined manually in MATLAB (MathWorks, Natick, USA) for machine learning; their locations were confirmed by a trained radiologist.
Automated Segmentation
Two separate U-Net models6 were implemented with alpha-balanced focal loss7 in Python 3.8 for segmentation on two ROIs in the multi-echo images: the entire prostate and PZ. Training of the U-Net was performed using n=144 slices (65%, 27 patients) and validated with n=36 slices (15%, 6 patients). Model evaluation was calculated on a test dataset of n=44 slices (20%, 7 patients) with three accuracy metrics averaged across the test dataset: pixel accuracy (sum of accurate pixels over the total number of pixels), Jaccard index8, and Dice coefficient8. The input to the model was the multi-echo T2 decay data of size 240x240x64. The U-Net model for the entire prostate zone had a learning rate of 10-4, a batch size of 2, 50 epochs, and gamma=4, and alpha=0.1. The U-Net model for the PZ had a learning rate of 7x10-4, a batch size of 15, 100 epochs, gamma=6, and alpha=0.1. The output of the model provided a binary mask of size 240x240 pixels.
Augmented T2W Imaging
The augmented image, IA, is defined as:
$$I_{A}=f_{rescale}\left(I_o*(1-w)+f_{norm}(I_o*f_W(I_{PW}))*W\right)$$
Where Io is the normalized multi-echo image (echo=4 to match the effective echo time of high-resolution T2W image) to values within the ROI, $$$w$$$ is a weighting scale from [0,1], and IPW is the standardized LWI parametric map in the ROI weighted by a function (fw). The normalization function (fnorm) maps all values between [0,1]. The rescaling function (frescale) maps values to the same relative minimum and maximum intensities in the ROI of Io. fw functions explored include a hyperbolic tangent and sigmoid function. $$$w$$$ values were swept between 0 and 1. Histograms of benign and cancerous pixel values in the ROI were generated with binning based on Sturge’s rule9. Two metrics to evaluate the efficacy of augmentation were histogram overlap between cancerous and benign pixels (OVL) and area under the curve (AUC) from receiver operating characteristics (ROC). ROC curves were calculated using normalized pixel intensity values as the probability for measuring malignancy within the PZ.
Results & Discussion
U-Net segmentation on the test dataset reports average Dice coefficients of 0.919 and 0.751 for the entire prostate and PZ segmentation, respectively. Segmentation metrics and an example segmentation of a single slice are shown in Table 1 and Figure 1. While PZ has lower segmentation accuracy, image augmentation is performed on the entire prostate and thus, minimally affects the purpose of augmentation.Table 2 shows the results of benign-to-malignant tissue separability of the original T2W image and augmented T2W with various weightings. Only LWF and gmT2 are reported as they showed the best separability from all LWI parameters and across the test dataset. The hyperbolic tangent function and a $$$w$$$ between 0.5 and 0.75 provided the best weighting on IPW overall. It is noted that OVL and AUC are susceptible to imaging artifacts and may not reflect cancer measurements in the pixels. However, these two metrics were effective in determining the best weighting parameters and functions. Figure 3 shows the effect of augmentation on both the multi-echo T2 and high-res T2W images. OVL is reported higher for GS 3+3 and lower for GS 3+4 compared with the un-augmented multi-echo T2 image. However, the augmented images both show better image contrast for cancer location and extent.
Conclusion
This work shows which weighting functions and parameters best augment T2W images to enhance PCa image contrast in the PZ. Future work includes expanding this process to the transition zone and exploring combinations of LWI parameters. In addition, the development of faster LWI protocols and at higher resolution will allow for clinical feasibility and better accuracy for radiological assessment of PCa.Acknowledgements
This research was supported by the Canadian Institutes for Health Research and the Natural Sciences and Engineering Research Council of Canada.
References
- Sabouri S, Chang SD, Savdie R, et al. Luminal Water Imaging: A New MR Imaging T2 Mapping Technique for Prostate Cancer Diagnosis. Radiology. 2017;284(2):451-459. doi:10.1148/radiol.2017161687
- Sabouri S, Fazli L, Chang SD, et al. MR measurement of luminal water in prostate gland: Quantitative correlation between MRI and histology. JMRI, 2017;46(3):861–869. doi:10.1002/jmri.25624
- Sabouri S, Chang SD, Goldenberg SL, et al. Comparing diagnostic accuracy of luminal water imaging with diffusion-weighted and dynamic contrast-enhanced MRI in prostate cancer: A quantitative MRI study. NMR Biomed, 2019l;32(2):e4048. doi:10.1002/nbm.4048
- Doucette J, Kames C, Rauscher A. DECAES - DEcomposition and Component Analysis of Exponential Signals. Z Med Phys, 2020;30(4):271–278. doi:10.1016/j.zemedi.2020.04.001
- Uribe CF, Jones EC, Chang SD, et al. In vivo 3T and ex vivo 7T diffusion tensor imaging of prostate cancer: Correlation with histology. Magn. Reason. Imaging, 2015;33(5), 577–583. doi:10.1016/j.mri.2015.02.022
- Ronnenberger O, Fischer P, Brox T. U-Net: Convolutional Networks for Biomedical Image Segmentation. Paper presented at: MICCAI 2015, Munich, Germany, 2015. doi:10.1007/978-3-319-24574-4_28
- Lin T, Goyal P, Girshick R, et al. Focal Loss for Dense Object Detection. Paper presented at: 2017 IEEE International Conference on Computer Vision (ICCV), Venice, Italy, 2017. doi:10.1109/ICCV.2017.324
- Bertels J, Eelbode T, Berman M, et al. Optimizing the Dice Score and Jaccard Index for Medical Image Segmentation: Theory & Practice. Paper presented at: MICCAI 2019, Shenzhen, China, 2019. doi:10.1007/978-3-030-32245-8_11
- Sturges HA. The Choice of a Class Interval. J Am Stat Assoc. 1926;21(153):65-66. doi:10.1080/01621459.1926.10502161
Figures
Table 2: Results of benign and cancerous pixel analysis from the peripheral zone of a single slice of a patient are shown. Two PCa Gleason scores (GS) are present: GS 3+3 and GS 4+3. Reported separability metrics are histogram overlap (OVL) and area-under-the-curve (AUC), with increase in separability from weighting in brackets. Lower OVL and high AUC refers to better separability. Highlighted in each column, for either parameter, is the best performing weighting function based on separability.
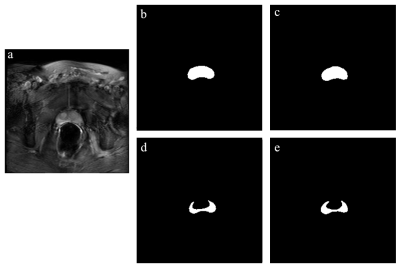
Figure 1: (a) shows the multi-echo image at echo=4. (b) and (d) are the ground truth regions of interest for the entire prostate and peripheral zone (PZ), respectively. (c) and (e) are the resultant segmented regions of the entire prostate and PZ by the trained U-Net model. The data presented is for a single slice from the test dataset.
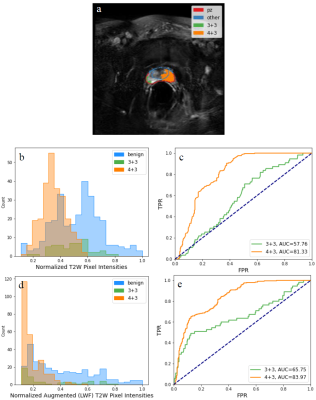
Figure 3: (a) shows the histologically examined slice for (b) the multi-echo T2 image (echo=4) and (d) the corresponding high-resolution (540x540) T2W image. (c) and (e) shows the augmented multi-echo T2 image and high-resolution T2W image, respectively, with LWF parameter. Weighting used $$$w$$$=0.75, and tanh weighting function. Mapping of weighted images to (e) was performed through a scaling deformation from 240x240 to a 540x540 FOV. Visually, the relative intensities are contrasted larger for both GS 3+3 and GS 4+3 in both PZ and non-PZ regions than in the T2W image.