0803
Open Science Initiative for Perfusion Imaging (OSIPI): DCE/DSC lexicon for reporting perfusion analysis pipelines1Division of Informatics, Imaging and Data Sciences, The University of Manchester, Manchester, United Kingdom, 2University of Sheffield, Sheffield, United Kingdom, 3the Netherlands Cancer Institute, Amsterdam, Netherlands, 4Genentech, Inc, San Francisco, CA, United States, 5Erasmus MC University Medical Center, Rotterdam, Netherlands, 6Harvard Medical School, Boston, MA, United States, 7University of Gothenburg, Gothenburg, Sweden, 8Karlsruhe Institute for Technology, Karlsruhe, Germany, 9University of Vienna, Vienna, Austria, 10Barrow Neurological Institute, Phoenix, AZ, United States, 11German Cancer Research Center DKFZ, Heidelberg, Germany, 12Mayo Clinic, Rochester, MN, United States, 13The University of Leeds, Leeds, United Kingdom
Synopsis
The Open Science Initiative for Perfusion Imaging (OSIPI) aims to improve the reproducibility of perfusion MRI research through creation of acquisition and analysis standards. Specifically, Task Force 4.2 was established to develop a lexicon of variables and processes to facilitate standardised reporting of dynamic contrast-enhanced (DCE-) and dynamic susceptibility contrast (DSC-) MRI analysis pipelines. Here we report progress towards these objectives by presenting the current DCE/DSC lexicon structure, and provide a use-case to encode a simple analysis pipeline.
Introduction
Dynamic contrast-enhanced (DCE) and dynamic susceptibility contrast (DSC) magnetic resonance imaging (MRI) are widely used in preclinical and clinical research studies to measure quantitative tissue perfusion properties[1,2]. Despite the best efforts of the research community, there is substantial variability in the nomenclature and manner in which analyses are reported, leading to inconsistencies between studies which ultimately hinders translation of DCE and DSC into routine clinical practice. The Open Science Initiative for Perfusion Imaging (OSIPI)[3] aims to eliminate the practice of duplicate development, improve the reproducibility of perfusion imaging research, and speed up the translation of perfusion imaging as tools for discovery science, drug development, and clinical practice. The aim of OSIPI Task Force 4.2[4] is to develop a standardised lexicon that enhances transparency and consistency when reporting DCE/DSC analyses. Here we describe the lexicon in its current state, and provide an example use-case showing how the lexicon can be used to encode a simple analysis pipeline.Methods
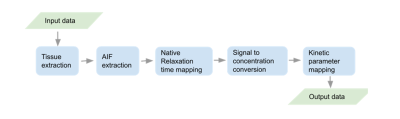
Task force (TF) 4.2 was formed in March 2020 and consists of experts in the fields of DCE- and DSC-MRI and DICOM standardization. Internal review of version 1. of the lexicon is due by Dec 2021 (milestone 4). Release of version 1.0 for public consultation is planned for Jan 2022 (milestone 5). Submission of version 1.0 for publication is planned for May 2022 (milestone 6). To demonstrate that the lexicon is now sufficiently comprehensive to encode pipelines from start to finish, an example encoding of a simple end-to-end pipeline (Figure 2) is presented.Results
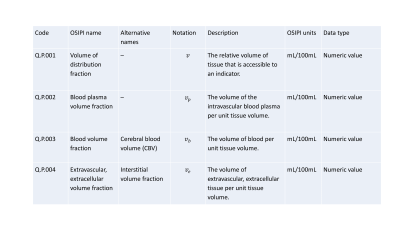
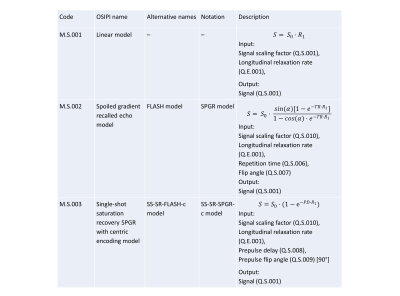
The lexicon scope is described here (https://docs.google.com/document/d/15aJmOhE4hyuK_mtWeEHjMs8kiKJ_R6y0lRZSXxQYTSw/edit). The lexicon currently includes 6 sections containing either quantities (170 items) or processes (44 general purpose processes, 42 perfusion models, 17 perfusion identities, 16 perfusion processes, and 3 pipelines) that are arranged in thematic groups (Figure 1). Each quantity or process is uniquely identified by a code and also assigned a human-readable name (Figure 3 and Figure 4). Codes are given in the format Section.Group.Entry (e.g. Q.P.001 refers to entry 001 from group P of section Q). Processes act as operators that generate output quantities based on given input quantities. They are semantic in nature, meaning they take inputs and return outputs without providing details of the specific algorithm used. Details of algorithms or implementations used in a specific analysis can be recorded by specifying an instance of the process (e.g. the GITHUB repo where the algorithm is stored). Figure 3 shows an excerpt from the Physiological quantities Group of the Perfusion Quantities Section. Figure 4 shows an excerpt from the MR signal models Group of the Perfusion Models Section. Figure 5 shows the simple analysis pipeline encoded using lexicon variables and processes.Discussion
We have presented the OSIPI DCE/DSC lexicon in its current form and reported the first application of its use to encode a simple end-to-end analysis pipeline. This demonstrates the lexicon is sufficiently complete to enable encoding of all basic inputs and outputs, and individual processes, required to convert image signal data into kinetic parameter maps. The content of the lexicon is intended to be dynamically growing - it is encouraged that the perfusion community should request new variables and processes to be added when necessary. Currently, lexicon sections are hosted on Google Docs, however by the time of publication we aim to publish lexicon sections as ‘data’ on GITHUB to enable issue tracking and auditable version control (by Summer 2022). Eventually we plan to develop an web-interface with a searchable database that enables pipelines to be designed and automatically converted into human-readable pseudocode and flow-charts for reporting purposes. The exact representation of pipeline encoding is still under development. The purpose here was not to present a polished pipeline representation, but to demonstrate that the lexicon content is sufficient to enable end-end description of a simple pipeline. In future, we will explore integration of the lexicon with existing computational pipeline languages such as WDL and CDL to maximise ease of use and uptake of the lexicon by the research community. In summary, we have presented the OSIPI DCE/DSC lexicon which we hope will help to standardize reporting of perfusion analyses and promote translation of DCE and DSC into routine clinical practice.Acknowledgements
No acknowledgement found.References
[1]. O'connor JP, Jackson A, Parker GJ, Roberts C, Jayson GC. Dynamic contrast-enhanced MRI in clinical trials of antivascular therapies. Nature reviews Clinical oncology. 2012 Mar;9(3):167-77.
[2]. Thrippleton MJ, Backes WH, Sourbron S, Ingrisch M, van Osch MJ, Dichgans M, Fazekas F, Ropele S, Frayne R, van Oostenbrugge RJ, Smith EE. Quantifying blood-brain barrier leakage in small vessel disease: review and consensus recommendations. Alzheimer's & Dementia. 2019 Jun 1;15(6):840-58.
[3] www.osipi.org
[4] www.osipi.org/task-force-4-2/
Figures