0782
Identification of Subcortical White Matter Biomarkers in Multiple Sclerosis Patients according to AVLT performance using Random Forest1Biomedical Imaging Center, Pontificia Universidad Católica de Chile, Santiago, Chile, 2Radiology Department, School of Medicine, Pontificia Universidad Católica de Chile, Santiago, Chile, 3Millennium Nucleus for Cardiovascular Magnetic Resonance, Santiago, Chile, 4Electrical Engineering Department, School of Engineering, Pontificia Universidad Católica de Chile, Santiago, Chile, 5Interdisciplinary Center of Neurosciences, Pontificia Universidad Católica de Chile, Santiago, Chile, 6Faculty of Engineering, Universidad de Concepción, Concepción, Chile, 7UNATI, Neurospin, CEA, Université Paris-Saclay, Gif-sur-Yvette, France, 8Neurology Department, School of Medicine, Pontificia Universidad Católica de Chile, Santiago, Chile, 9Neurology Service, Hospital Dr. Sótero del Río, Santiago, Chile
Synopsis
Radiological biomarkers of cognitive impairment in Multiple Sclerosis (MS) is still scarce. This study aimed to identify subcortical white matter biomarkers of cognitive impairment related to verbal episodic memory in MS patients and healthy controls, by using a random forest approach.
Introduction
Cognitive decline is recognized as a prevalent symptom of Multiple Sclerosis (MS), especially in episodic memory (1). These deficits are associated grey matter atrophy and white matter lesions in subcortical areas (2-4). Clinical tests detect mild cognitive changes in specific cognitive domains. Auditory Verbal Learning Test (AVLT) is performed as a screening tool to detect changes in verbal episodic memory (5). Current neurocognitive batteries may not identify early changes, precluding a timely diagnosis and treatment (6).MR techniques improve the understanding of the development of cognitive impairment in relapsing-remitting MS patients (RRMS) (7). Neuroimaging techniques, such as DTI, provide different indices to evaluate the axonal injury, demyelination, such as Fractional Anisotropy and Mean Axial, and Radial Diffusivity (FA,MD,AD,and RD,respectively) (8). Therefore, a biomarker that could detect patients with cognitive deficits, might benefit from early diagnosis and treatment.
Machine-learning (ML) algorithms have shown promising results in classifying MRI of patients with neurologic disorders (9,10). In particular, random forest (RF) uses a set of classification trees to estimates associations among different variables by permuting and bootstrapping the observations. Therefore, the RF method has high prediction accuracy compared to other classification and regression algorithms (11).
In this study, we used a ML approach, to identify subcortical white matter biomarkers between healthy controls (HC) and RRMS patients with or without cognitive impairment in verbal episodic memory, as determined by the results from the AVLT test, with RF.
Materials and Methods
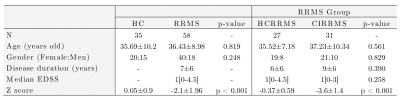
Diffusion-weighted and T1-weighted images were acquired in a 3T MRI scanner (Philips Ingenia, Best, Netherlands) in 35 HC (57% female) and 58 RRMS patients (69% female). The local ethics committee approved the study, and RRMS patients were diagnosed according to 2017 McDonald's criteria (7). Table 1 summarizes the demographic data. All patients were evaluated with an expanded disability status scale (EDDS) and verbal memory assessment using AVLTVIII (5). All test scores were normalized in Z-scores. Using the Z-score, patients and healthy controls were categorized as HC and HCRRMS with a Z score>= -1.5, and CIRRMS with a Z score< -1.5.Diffusion-weighted images were processed to obtain FA, AD, MD, and RD maps using diffusion tensor imaging (DTI). T1-weighted images were used as an anatomical reference. All preprocessing steps were performed in SPM12 (12). We used the LNAO-SWM79 U-fiber Atlas as a mask to obtain the mean FA, AD, MD, and RD map to each subject's U-fiber (13).
Furthermore, a ML model was designed to select U-fiber maps that adequately separate healthy controls (HC) vs. HCRRMS, HCRRMS vs. CIRRMS, HC vs. CIRRMS, and between the three classes. We used sequential selection forward (SFS) and principal component analysis (PCA) as feature selection algorithms to eliminate highly correlated or constant features that maximized accuracy (14). We use RF as a classifier. Besides that, we used bootstrap-aggregated decision trees to combine the results of many decision trees, which reduces the effects of overfitting and improves generalization. Therefore, at each split/node, a variable is selected to maximize the variance explanation of the dependent variable. In each round of training, 1000 decision trees were generated with a maximum allowed tree depth of five (15). Analysis was performed using 10-fold stratified cross-validation. Confusion matrices were obtained from the results in each training and validation sample. The accuracy, sensitivity, specificity, and precision of the search strategies were calculated (16).
The algorithms were implemented in MATLAB using Balu (17) and the Statistics and Machine Learning MATLAB Toolbox.
Results
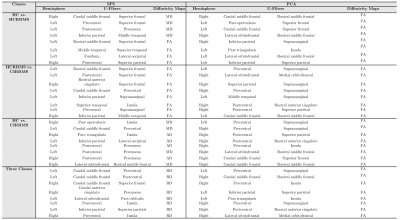
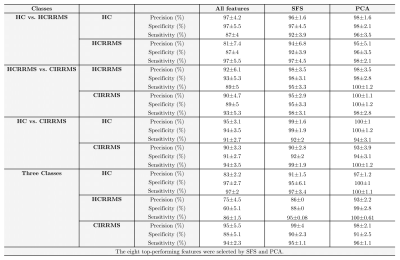
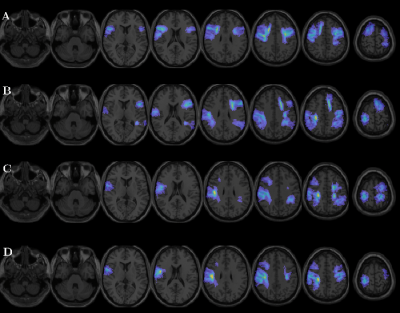
From the U-fibers maps obtained, the eight features that best differentiated the different combinations of classes were selected using both SFS and PCA. Table 2 shows the U-fibers maps features, selected SFS, and PCA. In SFS, the eight variables presenting with the lowest separability and higher accuracy once used the RF were retained for the HC vs. HCRRMS. They correspond to FA and MD U-fiber maps, located in frontal, temporal, and parietal lobes. In PCA, the eight top-performing features correspond to FA U-fiber maps, located in frontal, temporal, and parietal lobes for classifying HC vs. HCRRMS.The results of the RF analysis in the different subgroups of subjects are reported in Table 3. Using features selected by SFS, RF resulted in an accuracy of over 92%. Using features selected by PCA, almost all combinations of classes were close to 97% accuracy. The best results were obtained by combining eight features selected by PCA using the RF. Table 3 shows the precision, sensibility, and sensibility with the best performance. In the different groups of classes, the RF analysis showed heterogeneous performance. Low performances were observed when the design was unbalanced. Finally, Figure 1 shows the U-fibers selected by PCA with the best accuracy performance in different combinations of classes.
Conclusions
We showed that the RF classification robustly and automatically discriminates between HC and MS patients with or without verbal episodic memory impairment as determined by the AVLTVIII, setting up 97% accuracy. Furthermore, we used PCA as a feature selection algorithm to determine eight U-fibers features that can characterize RRMS patients. The approach allows identifying areas related to lower performance in the AVLTVII. The identification and assessment of these areas may help to early identify signs of early cognitive impairment in MS.Acknowledgements
This work has been funded by projects PIA-ACT192064, the Millennium Nucleus on Cardiovascular Magnetic Resonance NCN17_129 of the Millennium Science Initiative, and Fondecyt 1181057, from the National Agency for Research and Development, ANID.References
1. Benedict RHB, Amato MP, DeLuca J, et al. Cognitive impairment in multiple sclerosis: clinical management, MRI, and therapeutic avenues. Lancet Neurol. 2020 Oct;19(10):860-871. doi: 10.1016/S1474-4422(20)30277-5. Epub 2020 Sep 16. PMID: 32949546.
2. Brownell B, Hughes JT. The distribution of plaques in the cerebrum in multiple sclerosis. J Neurol Neurosurg Psychiatry. 1962;25:315–20
3. Miki Y, Grossman RI, Udupa JK, et al. Isolated U-fiber involvement in MS: preliminary observations. Neurology. 1998;50(5): 1301–6.
4. Lazeron RH, Langdon DW, Filippi M, et al. Neuropsychological impairment in multiple sclerosis patients: the role of (juxta) cortical lesion on FLAIR. Mult Scler. 2000;6(4): 280–5.
5. Bender HA, Cole JR, Aponte-Samalot M, Cruz-Laureano D, Myers L, Vazquez BR, Barr WB. Construct validity of the Neuropsychological Screening Battery for Hispanics (NeSBHIS) in a neurological sample. J Int Neuropsychol Soc. 2009 Mar;15(2):217-24. doi: 10.1017/S1355617709090250. Epub 2009 Feb 12. PMID: 19215638.
6. Sumowski JF, Benedict R, Enzinger C, et al. Cognition in multiple sclerosis: State of the field and priorities for the future. Neurology. 2018 Feb 6;90(6):278-288. doi: 10.1212/WNL.0000000000004977. Epub 2018 Jan 17. PMID: 29343470; PMCID: PMC5818015.
7. Thompson AJ, Banwell BL, Barkhof F, et al. Diagnosis of multiple sclerosis: 2017 revisions of the McDonald criteria. Lancet Neurol. 2018 Feb;17(2):162-173. doi: 10.1016/S1474-4422(17)30470-2. Epub 2017 Dec 21. PMID: 29275977.
8. Sbardella E, Tona F, Petsas N, et al. DTI Measurements in Multiple Sclerosis: Evaluation of Brain Damage and Clinical Implications. Mult Scler Int. 2013;2013:671730.
9. Klöppel S, Stonnington CM, Chu C, et al. Automatic classification of MR scans in Alzheimer’s disease. Brain 2008;131:681–689.
10. Wottschel V, Alexander DC, Kwok PP, et al. Predicting outcome in clinically isolated syndrome using machine learning. NeuroImage Clin 2015;7:281–287.
11. Verikas A, Gelzinis A, Bacauskiene M: Mining data with random forests: asurvey and results of new tests. Pattern Recognition 2011, 44(2):330–349.
12. Montalba C, Labbe, T, Andia M, et al. Evaluation of PASAT test performance and diffusivity indices in U-fiber regions in healthy subjects and RRMS patients. 10. Proc. Intl. Soc. Mag. Reson. Med. 29 (2021).
13. Guevara M, Román C, Houenou J, et al. Reproducibility of superficial white matter tracts using diffusion-weighted imaging tractography. Neuroimage. 2017 Feb 15;147:703-725.
14.- Mery D. Computer Vision for X-Ray Testing: Imaging, Systems, Image Databases, and Algorithms, Springer (1st ed) 2015
15.- Ciaburro G. MATLAB for Machine Learning: Practical examples of regression, clustering and neural networks, Publisher: Packt Publishing, 2017, ISBN: 978-1788398435.
16.- Witten I and Frank E, Data mining: practical machine learning tolls and techniques, San Mateo (2nd ed.), 2005.
17.- Mery D. BALU: A Matlab toolbox for computer vision, pattern recognition and image processing. 2011 http://dmery.ing.puc.cl/index.php/balu.
Figures