0778
“Better together”: a combination of machine learning and radiomics to predict long-term disability in Multiple Sclerosis1University of Naples Federico II, Naples, Italy, 2National Research Council, Naples, Italy
Synopsis
The identification of early biomarkers to predict the disability accumulation is crucial in Multiple Sclerosis (MS). We performed a combined radiomics and Machine Learning (ML) study to predict long-term clinical changes in MS. Radiomics data were extracted from data of 177 patients with a 10-years clinical follow-up available. The model based on the recursive elimination of the features combined with the Extra Trees classifier was able to obtain a maximum precision for each endpoint of 0.71 and 0.69 for cognitive and motor disability, respectively.This combined radiomics-ML approach seems to be a feasible tool for long-term clinical prediction in MS.
Introduction
Clinical progression and disability accumulation are heterogeneous phenomena inevitably occurring in patients with multiple sclerosis (MS). The identification of potential early biomarkers capable of predicting the occurrence of these phenomena is crucial, and an actual challenge to researchers working in this field 1. Among all the different biomarkers available in MS, MRI plays a pivotal role, being routinely performed in these patients to monitor the disease and identify pathophysiological changes related to the disease 2. Although several metrics have been suggested as potential biomarkers able to predict the clinical status of these patients, information about MRI long-term predictors of clinical disability are still relatively lacking. Based on volumetric and connectivity features extracted from MRI, we aimed to detect the accuracy of Machine Learning (ML) models to predict long-term clinical changes in MS patients, namely cognitive impairment (CI) and confirmed disability progression (CDP).Methods
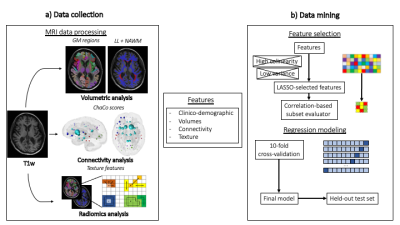
This retrospective evaluation was based on 177 MS patients’ data, who underwent a 3T brain MRI scan, and had a long-term (around 10 years) clinical follow-up available.Motor function assessment was performed via EDSS at baseline and follow-up, while cognitive functions were evaluated via BICAMS at follow-up. MRI protocol included a FLAIR sequence and a 3D T1-volume. Disconnection scores (inferred from the position and load of the white matter lesions defined on the FLAIR sequence) and radiomic characteristics from the atlas 116 regions of gray matter defined by the Automated Anatomical Atlas (AAL) 3 were extracted. Feature extraction was carried out using PyRadiomics v3.0.1 4.Machine learning analyses were performed using scikit-learn Python package 5. Extra Trees, Logistic Regression and Support Vector Machine are the three ML algorithms used to predict both CI and CDP. A 10-fold cross-validation in the training set (n=132) was used to optimize the algorithms with final model evaluation on the held-out test set (n=45).
Results
A total of 127549 features are stable out of the 127764 that were extracted. After the low variance (n = 484) and highly interrelated (= 100860) parameters were removed, LASSO feature selection identified a subset of 19 and 13 features, all but 2 from radiomics, for CI and CDP estimation, respectively. The final feature set for each model was obtained through recursive feature elimination (RFE). Regarding the CI class prediction analysis, the RFE identified a 16-, 17- and 19-feature subsets, with an accuracy on the test set of 0.69, 0.64 and 0.62 for the Extra Trees, Logistic Regression and Support Vector Machine classifiers, respectively. Similarly, in relation to the CDP prediction assessment, the RFE selected a 13-, 12- and 10-feature subsets with a 0.71, 0.69 and 0.67 accuracy values on the test set, respectively (Figure 1).Discussion
In this study we were able to demonstrate the feasibility of a combined approach that, integrating ML and textural features extracted from routine brain MRI scans, was able to predict long-term disability in MS patients.The search of biomarkers able to predict the progression of the disease is one of the main area of research in MS. Nevertheless, only the recent availability of large datasets have permitted this growing implementation of ML in research settings witnessed in the last years in the field of neuroimaging. The integration of such analyses with information extracted from signal intensity patterns, assessed by quantitative texture analysis, has proven to be able in obtaining meaningful information from a clinical standpoint 6.
Although promising, the approach here proposed shows some limitations, including the relatively low number of cases included, or the known reported instability of radiomics features due to variations in scanner and images, limiting at the moment the generalizability of our models across different sequences and scanners, although paving the road to future multicentric studies.
Conclusion
The evaluation of radiomic features using a ML analysis proves to be a feasible and promising tool for long-term prediction of both physical and cognitive disability in MS patients.Acknowledgements
No acknowledgement found.References
1. Pinto MF, Oliveira H, Batista S, Cruz L, Pinto M, Correia I, Martins P, Teixeira C. Prediction of disease progression and outcomes in multiple sclerosis with machine learning. Sci Rep. 2020 Dec 3;10(1):21038. doi: 10.1038/s41598-020-78212-6. PMID: 33273676; PMCID: PMC7713436
2. Wattjes MP, Ciccarelli O, Reich DS, Banwell B, de Stefano N, Enzinger C, Fazekas F, Filippi M, Frederiksen J, Gasperini C, Hacohen Y, Kappos L, Li DKB, Mankad K, Montalban X, Newsome SD, Oh J, Palace J, Rocca MA, Sastre-Garriga J, Tintoré M, Traboulsee A, Vrenken H, Yousry T, Barkhof F, Rovira À; Magnetic Resonance Imaging in Multiple Sclerosis study group; Consortium of Multiple Sclerosis Centres; North American Imaging in Multiple Sclerosis Cooperative MRI guidelines working group. 2021 MAGNIMS-CMSC-NAIMS consensus recommendations on the use of MRI in patients with multiple sclerosis. Lancet Neurol. 2021 Aug;20(8):653-670. doi: 10.1016/S1474-4422(21)00095-8. Epub 2021 Jun 14. PMID: 34139157
3. Tzourio-Mazoyer N, Landeau B, Papathanassiou D, Crivello F, Etard O, Delcroix N, Mazoyer B, Joliot M. Automated anatomical labeling of activations in SPM using a macroscopic anatomical parcellation of the MNI MRI single-subject brain. Neuroimage. 2002 Jan;15(1):273-89. doi: 10.1006/nimg.2001.0978. PMID: 11771995
4. van Griethuysen JJM, Fedorov A, Parmar C, Hosny A, Aucoin N, Narayan V, Beets-Tan RGH, Fillion-Robin JC, Pieper S, Aerts HJWL. Computational Radiomics System to Decode the Radiographic Phenotype. Cancer Res. 2017 Nov 1;77(21):e104-e107. doi: 10.1158/0008-5472.CAN-17-0339. PMID: 29092951; PMCID: PMC5672828
5. Pedregosa F, Varoquaux G, Gramfort A, et al. Scikit-learn: Machine Learning in Python. J Mach Learn Res 2011;12:2825–2830
6. Pontillo G, Tommasin S, Cuocolo R, Petracca M, Petsas N, Ugga L, Carotenuto A, Pozzilli C, Iodice R, Lanzillo R, Quarantelli M, Brescia Morra V, Tedeschi E, Pantano P, Cocozza S. A Combined Radiomics and Machine Learning Approach to Overcome the Clinicoradiologic Paradox in Multiple Sclerosis. AJNR Am J Neuroradiol. 2021 Sep 16. doi: 10.3174/ajnr.A7274. Epub ahead of print. PMID: 34531195
Figures
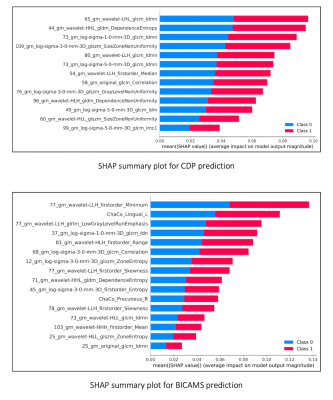
Figure 1
SHAP summary plots showing features predictive of CDP and CI in MS patients, calculated from EDSS changes and follow-up BICAMS scores respectively. On the x-axis mean SHAP values are reported, while notations on y-axis reflect the selected features of the corresponding gray matter region.
SHAP: Shapely Additive Explanation; CDP: Confirmed Disability Progression; CI: Cognitive Impairment; MS: Multiple Sclerosis; EDSS: Expanded Disability Status Scale; Class 0: not progressed/not impaired; Class 1: progressed/impaired.