0681
ImUnity: a generalizable VAE-GAN solution for multicenter MR image harmonization1Grenoble Institut of Neurosciences, Grenoble, France
Synopsis
ImUnity is an original deep-learning model designed for efficient and flexible MRI harmonization. A VAE-GAN network is coupled with a confusion and a biological preservation module. It ‘corrects’ MR images that can be used for various multi-center studies. Using 3 open source databases, we show that ImUnity: outperforms state-of-the-art methods in terms of quality of images generated; removes sites/scanner bias while improving patients classification; harmonizes data coming from new sites/scanners and allows the selection of multiple MR reconstructed images according to the desired applications. Tested on T1w images, ImUnity could be generalized to other types of medical images.
Introduction
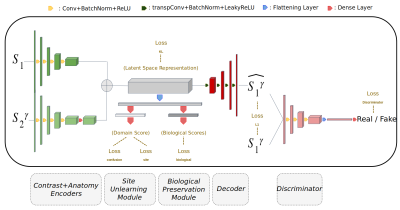
Pooling images from multi-center MR studies in order to approach a particular clinical question does not guarantee an increase in statistical power because of a parallel increase in non-biological variance (due to variations in hardware or sequence parameters). Many harmonization methods have been developed over the last few years to tackle this issue 1,2,3. However, existing solutions have the downsides of requiring travelling subjects (scanned at every site), special combination of MR sequences or new training of the algorithm every time new sites are added to the database.We propose here a new method that can offer a fast and flexible harmonization solution able to correct inter-sites/scanners contrast variations while preserving biological information. Our self-supervised Variational AutoEncoder (VAE-GAN) architecture uses multiple slices from the same individual and randomized image contrast transformations for its training. It also unlearns center bias using a confusion module connected to its bottleneck while a biological module can ensure that clinical features are preserved in the latent space. This architecture allows unseen data to be harmonized without the need for fine-tuning, and proposes multiple target sites to allow users to choose multiple MR image reconstructions. We evaluated our approach using 3 open access databases and ran multiple experiments to test the quality of reconstructed images, the capacity to remove site or scanner information and to improve patient classification after harmonization.Materials and methods
The model’s architecture is shown in Fig-1. We used 3 open access databases for training and validation: (A) ABIDEII4: a multi-center database focusing on Autism Spectrum Disorder (ASD)(n=621 T1w scans were used). (B) OASIS35: a travelling subjects database gathering n=769 T1w scans from subjects who underwent 4 MR sessions using different scanners from the same site. (C) SRPBS6: a database focusing on adult travelling subjects between different sites (9 subjects scanned in 11 different sites on 3 different scanners). Multiple experiments were run to evaluate different aspects of the model:Experiment 1. Impact on image quality in multi-sites or multi-scanner harmonization was assessed using data from traveling subjects (ground truth). We computed Structural Similarity Index Measure (SSIM) between different acquisitions for each patient before and after harmonization. An arbitrary reference domain was chosen for harmonization.
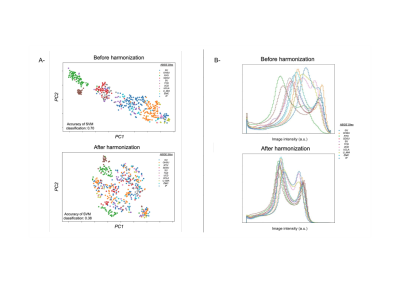
Experiment 2. Ability to remove site information was assessed using tSNE and standard SVM classification before and after harmonization of the ABIDE database..
Experiment 3. Improvement in autism disorder prediction in children from the ABIDE database was assessed using a support vector machine (SVM) classification before and after harmonization.
To demonstrate the flexibility of ImUnity, all experiments were performed with the same model trained on the ABIDE database, unless specified. OASIS and SRPBS were used for the validation parts only.
Models were trained using an Nvidia GeForce 2080 RTX during 2000 epochs using a learning rate of 10E-4 and Adam optimizer.
Results
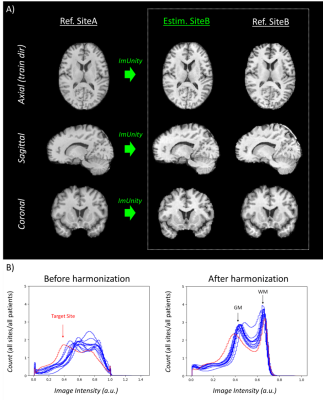
Experiment 1: Fig-2-A shows the results obtained in one traveling subject from the SRPBS database. We can observe contrast differences before harmonization and homogenization after ImUnity. It is interesting to observe that the anatomical structures of the input contrast reference are not propagated through the model, a crucial point as we want to preserve the anatomical information. Fig-2-B shows the effects of ImUnity’s harmonization on image intensity distributions for all selected subjects from the SRPBS database. The model was used to harmonize every image to a target site. An alignment of histograms can be clearly observed after harmonization, with both gray and white matter peaks shifted. Fig-3 presents ImUnity’s impact on SSIM metrics. ImUnity increases SSIM in all cases and provides better results compared to other deep learning approaches. Moreover, results from multi-scanner harmonization show that ImUnity performs well independently of the chosen reference domain.Experiment 2: Fig-4 depicts ImUnity’s ability to remove site effects on the ABIDE dataset. Before harmonization, the presence of site clusters is clear. Once the data are harmonized, the points are shuffled and the accuracy of the SVM site prediction decreases. Intensity histograms between sites are also more homogenous after harmonization. This confirms the removal of site biases as the classifier is no longer able to correctly separate the sites and as intensity distributions have been realigned.
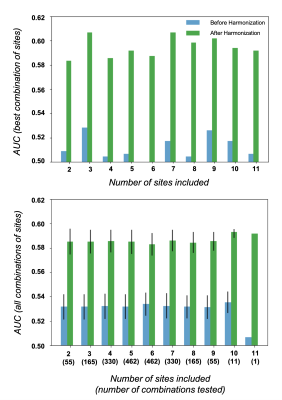
Experiment 3: Fig-5 shows the capacity of our harmonization tool to improve ASD prediction from the ABIDE dataset (anatomical data only). We used the same trained model to test the influence of different numbers of sites included in the database as well as different combinations of those sites. In every case, we observed an increase of the classifier’s AUC and accuracy after harmonization.
Conclusion
The proposed approach ensures realistic outputs, allows removal of idiosyncratic datasets bias and the preservation of biological information. Our results show that ImUnity reaches state-of-the-art image quality in traveling patients and improves ASD classification. Requiring only one type of MR sequence without the need of matching subjects, it can be generalized to unseen sites and can be used to harmonize MR images to different reference domains without a new training phase. We are currently working on improving ImUnity with a 2.5D2 approach to improve output realism and with a new architecture that allows simultaneous multi-sequence harmonization.Acknowledgements
Stenzel Cackowski is supported by MIAI@Grenoble Alpes (ANR 19-P3IA-003).References
- Fortin, J.P., et al., 2017, Harmonization of multi-site diffusion tensor imaging data. Neuroimage 161, 149–170. URL: https://www.ncbi.nlm.nih.gov/pmc/articles/PMC5736019/, doi: 10.1016/j.neuroimage.2017.08.047
- Dewey, B.E., et al., 2019, DeepHarmony: A deep learning approach to contrast harmonization across scanner changes. Magn. Reason. Imaging, doi: 10.1016/j.mri.2019.05.041
- Zuo et al., 2021, Unsupervised MR harmonization by learning disentangled representations using information bottleneck theory. Neuroimage 243, doi: 10.1016/j.neuroimage.2021.118569
- Di Martino, et al., 2014. The autism brain imaging data exchange: towards a large-scale evaluation of the intrinsic brain architecture in autism. Mol. Psychiatry 19, 659–667, doi: 10.1038/mp.2013.78
- LaMontagne, et al., 2019. OASIS-3: Longitudinal Neuroimaging, Clinical, and Cognitive Dataset for Normal Aging and Alzheimer Disease. URL: https://www.medrxiv.org/content/10.1101/2019.12.13.19014902v1, doi: 10.1101/2019.12.13.19014902
- Tanaka, et al., 2021. A multi-site, multi-disorder resting-state magnetic resonance image database. Sci Data 8, 227. URL: http://www.nature.com/articles/s41597-021-01004-8, doi: 10.1038/s41597-021-01004-8
Figures
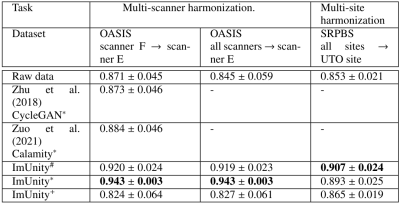
SSIM in traveling subjects for multi-scanner (OASIS database) and multi-site (SRPBS database) harmonization. Results are compared to the literature when available.
*: Model trained on OASIS database (n=1072);
#: Model trained on ABIDE database (n=545);
+: Model trained on SRPBS database (n=81)