0552
Cycle-Consistent Adversarial Transformers for Unpaired MR Image Translation1Department of Electrical and Electronics Engineering, Bilkent University, Ankara, Turkey, 2National Magnetic Resonance Research Center (UMRAM), Bilkent University, Ankara, Turkey, 3Electrical Engineering, Stanford University, Stanford, CA, United States, 4Neuroscience Program, Aysel Sabuncu Brain Research Center, Bilkent University, Ankara, Turkey
Synopsis
Translating acquired sequences to missing ones in multi-contrast MRI protocols can dramatically reduce scan costs. Neural network models devised for this purpose are characteristically trained on paired datasets, which can be difficult to compile. Moreover, these models exclusively rely on convolutional operators with undesirable biases towards feature locality and spatial invariance. Here, we present a cycle-consistent translation model, ResViT, to enable training on unpaired datasets. ResViT combines localization power of convolution operators with contextual sensitivity of transformers. Demonstrations on multi-contrast MRI datasets indicate the superiority of ResViT against state-of-the-art translation models.
Introduction
Multi-contrast Magnetic Resonance Imaging (MRI) is a key technology that is often underutilized due to costs associated with multiple separate scans. Thus, the ability to translate available source contrasts to unacquired target contrasts has important practical implications1,2,3. Learning-based translation methods are commonly trained on spatially-registered source-target images from matching subjects using pixel-wise error terms4,5. Yet, it is often challenging to compile large datasets of paired images, raising interest in methods that can learn from unpaired data6,7,8,9,10.State-of-the-art methods for unpaired image translation are pervasively based on cycle-consistent adversarial architectures with convolutional backbones5,9. However, convolutional networks with compact filters shared across the entire image exhibit a strong bias towards feature locality and spatial invariance. This inductive bias hampers learning of long-range spatial dependencies that can be critical for medical image translation11. Emergent transformer models in computer vision instead leverage self-attention mechanisms that forego inductive biases to enhance learning of global context12,13. Here, we introduce a novel deep learning model for unpaired medical image translation, cycle-consistent residual vision transformer (ResViT), that combines the sensitivity of vision transformers to context, localization power of convolutional networks, and realism of adversarial learning.
Methods
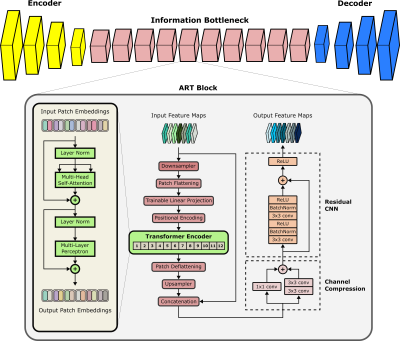
ResViT Model: ResViT is an adversarial architecture containing generator and discriminator subnetworks. The generator follows an encoder-decoder pipeline with a central information bottleneck, whereas the discriminator is a PatchGAN model14.a) Encoder: The encoder comprises a cascade of strided convolutional layers to extract intermediate-level image features.
b) Information Bottleneck: The information bottleneck contains a cascade of novel aggregated residual transformer (ART) blocks to extract local and global representations. The primary function of ART blocks is to aggregate the information distilled via residual convolutional and transformer branches (Figure 1). ART blocks are designed to perform the following operations:
- Due to computational constraints, received feature maps downsampled spatially with a stack of strided convolutions.
- Downsampled maps are split into non-overlapping patches and flattened.
- Flattened patches ($$$f_j^i$$$) are embedded onto an $$$N_D$$$-dimensional space via learnable linear projections($$$P_E$$$) and learnable positional encodings($$$P_E^{pos}$$$):
$$z_0 = [f_j^1P_E;f_j^2P_E;\dots;f_j^{N_P}P_E]+P_E^{pos}$$ where $$$z_0$$$ denotes patch embeddings. - The transformer block processes patch embeddings via 12 layers of multi-head self-attention and multi-layer perceptrons.
- Transformer output is deflattened and upsampled to match the size of input feature maps.
- Upsampled map is then fused with the input feature map, followed by channel compression.
- Compressed maps are processed via a residual convolutional network (CNN) to distill learned structural and contextual representations.
Cycle-consistent ResViT: The bidirectional mappings in the proposed cycle-consistent translation method (($$$x_2 \rightarrow x_1$$$, $$$x_1 \rightarrow x_2$$$), where $$$x_1$$$ and $$$x_2$$$ are the source and target contrasts) are implemented via two ResViT models with generators ($$$G_1$$$, $$$G_2$$$) and discriminators ($$$D_1$$$, $$$D_2$$$). To train these sub-networks on unpaired data, a cycle-consistency loss is used where a source or target image is remapped onto itself via the bidirectional mappings (Figure 2):
$$\mathcal{L}_{cyc} = \mathrm{E}[||x_1 - G_1(G_2(x_1))||_1] + \mathrm{E}[||x_2 - G_2(G_1(x_2))||_1]$$ In addition, an adversarial loss is incorporated to enhance capture of high-frequency details:
$$\mathcal{L}_{adv} = - \mathrm{E}[(D_1(x_1) - 1)^2] - \mathrm{E}[D_1(G_1(x_2))^2] - \\\mathrm{E}[(D_2(x_2) - 1)^2] - \mathrm{E}[D_2(G_2(x_1))^2]$$The overall optimization objective is expressed as:
$$\mathcal{L}_{ResViT} = \lambda_{cyc}\mathcal{L}_{cyc}+\lambda_{adv}\mathcal{L}_{adv}$$ where $$$\lambda_{cyc}$$$ and $$$\lambda_{adv}$$$ denotes the relative weightings for cycle-consistency and adversarial losses, respectively.
Modeling Procedures: Model training was continued for 200 epochs with Adam16 with $$$\beta_1=0.5,\,\beta_2=0.999$$$ with learning rate $$$0.0002$$$. $$$\lambda_{cyc}, \lambda_{adv}$$$ were set to 100,1 based on cross-validation. No spatial registration was performed between source-target images, which were sampled randomly to form unpaired sets of training/testing samples.
Dataset: We demonstrated the proposed ResViT model on the BRATS16 dataset. T1-weighted, T2-weighted brain MR images from 55 subjects were analyzed. 25, 10, 20 subjects were reserved for training, validation, and testing, respectively. From each subject, 100 axial cross-sections containing brain tissues were selected.
Results
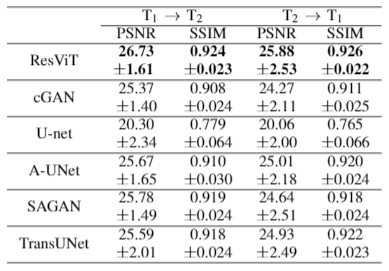
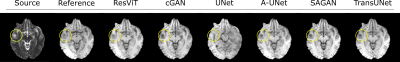
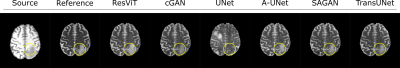
We demonstrated the proposed ResViT model for unpaired multi-contrast MRI translation against several state-of-the-art models, including convolutional (cGAN5, U-net17), attention-augmented convolutional (Attention U-Net18, SAGAN19), and transformer-based (TransUNet12) architectures. All methods were trained in a cycle-consistent and adversarial setup. We considered $$$T_1\rightarrow T_2$$$ and $$$T_2\rightarrow T_1$$$ mappings. PSNR and SSIM metrics for all methods are listed in Figure 3. ResViT achieves the highest performance in both tasks. On average across tasks, ResViT outperforms convolutional GANs by 3.90 dB PSNR and 0.85% SSIM, attention-based GANs by 1.40 dB PSNR and 1.45% SSIM, and TransUNet by 2.10 dB PSNR and 1.26% SSIM. Representative target images from all methods are displayed in Figures 4,5. ResViT improves translation performance in regions that are depicted sub-optimally in competing methods. Importantly, ResViT reliably captures pathology in patients in contrast to competing methods with inaccurate depictions.Conclusion
In this study, we proposed a novel adversarial model to overcome the limitations of traditional cycle GANs, which often fail to capture long-range spatial context11,12. ResViT instead aggregates convolutional and attention-based features to preserve local precision and contextual sensitivity. We comprehensively demonstrated ResViT in unpaired translation tasks in multi-contrast MRI. Our results indicate that ResViT outperforms convolutional GANs with or without attention mechanisms as well as a recent transformer architecture. Therefore, ResViT is a promising candidate for medical imaging tasks requiring high sensitivity to local and global context.Acknowledgements
This study was supported in part by a TUBITAK BIDEB, a TUBA GEBIP 2015 fellowship, a BAGEP 2017 fellowship.References
1. J. Denck, J. Guehring, A. Maier, and E. Rothgang, “Enhanced magnetic resonance image synthesis with contrast-aware generative adversarial networks,” Journal of Imaging, vol. 7, p. 133, 08 2021.
2. Wang G, Gong E, Banerjee S, Martin D, Tong E, Choi J, Chen H, Wintermark M, Pauly JM, Zaharchuk G. Synthesize High-Quality Multi-Contrast Magnetic Resonance Imaging From Multi-Echo Acquisition Using Multi-Task Deep Generative Model. IEEE Trans Med Imaging. 2020 Oct;39(10):3089-3099.
3. Lee, D., Moon, WJ. & Ye, J.C. Assessing the importance of magnetic resonance contrasts using collaborative generative adversarial networks. Nat Mach Intell 2, 34–42 (2020)
4. Kim S, Jang H, Hong S, Hong YS, Bae WC, Kim S, Hwang D. Fat-saturated image generation from multi-contrast MRIs using generative adversarial networks with Bloch equation-based autoencoder regularization. Med Image Anal. 2021 Oct;73:102198.
5. S. U. Dar, M. Yurt, L. Karacan, A. Erdem, E. Erdem, and T. Çukur, “Image synthesis in multi-contrast MRI with conditional generative adversarial networks,” IEEE Transactions on Medical Imaging, vol. 38, no. 10, pp. 2375–2388, 2019.
6.J.-Y. Zhu, T. Park, P. Isola, and A. A. Efros, “Unpaired image-to-image translation using cycle-consistent adversarial networks,” 2017 IEEE International Conference on Computer Vision (ICCV), pp. 2242–2251, 2017.
7. Pan, Y., Liu, M., Lian, C., Zhou, T., Xia, Y., & Shen, D. (2018). Synthesizing Missing PET from MRI with Cycle-consistent Generative Adversarial Networks for Alzheimer's Disease Diagnosis. Medical image computing and computer-assisted intervention : MICCAI International Conference on Medical Image Computing and Computer-Assisted Intervention, 11072, 455-463
8. Chartsias, A., Joyce, T., Dharmakumar, R., Tsaftaris, S.A., 2017. Adversarial image synthesis for unpaired multi-modal cardiac data, in: Simulation and Synthesis in Medical Imaging. pp. 3–13.
9. H. Yang et al., "Unsupervised MR-to-CT Synthesis Using Structure-Constrained CycleGAN," in IEEE Transactions on Medical Imaging, vol. 39, no. 12, pp. 4249-4261, Dec. 2020, doi: 10.1109/TMI.2020.3015379.
10. Y. Hiasa, Y. Otake, M. Takao, T. Matsuoka, K. Takashima, J. Prince, N. Sugano, and Y. Sato, “Cross-modality image synthesis from unpaired data using CycleGAN: Effects of gradient consistency loss and training data size,” ArXiv, abs/1803.06629, 2018.
11. Dalmaz, O., Yurt, M., & Çukur, T. ResViT: Residual vision transformers for multi-modal medical image synthesis. ArXiv, abs/2106.16031, 2021
12. J. Chen, Y. Lu, Q. Yu, X. Luo, E. Adeli, Y. Wang, L. Lu, A. L. Yuille, and Y. Zhou, “Transunet: Transformers make strong encoders for medical image segmentation,” ArXiv, vol. abs/2102.04306,2021.
13. A. Dosovitskiy, L. Beyer, A. Kolesnikov, D. Weissenborn, X. Zhai, T. Unterthiner, M. Dehghani, M. Minderer, G. Heigold, S. Gelly, J. Uszkoreit, andN. Houlsby, “An image is worth 16x16 words: Transformers for image recognition at scale,” ArXiv,vol. abs/2010.11929, 2021.
14. P. Isola, J .-Y. Zhu, T. Zhou, and A. A. Efros, “Image-to-image translation with conditional adversarial networks,” CVPR, 201 7.
15. D. P. Kingma and J. Ba, “Adam: A method for stochastic optimization,” in3rd International Conference on Learning Representations, ICLR 2015, San Diego, CA, USA, May 7-9, 2015, Conference Track Proceedings(Y. Bengio and Y. LeCun, eds.), 2015.
16. B. H. Menze, A. Jakab, S. Bauer, J. Kalpathy-Cramer, and et al., “The multimodal brain tumor image segmentation benchmark (brats),”IEEE transactions on Medical Imaging, vol. 34, no. 10, pp. 1993–2024, 2015.
17. O. Ronneberger, P. Fischer, and T. Brox, “U-Net: Convolutional networks for biomedical image segmentation,”Medical Image Computing and computer-Assisted Intervention – MICCAI 2015, May 2015.
18. O. Oktay, J. Schlemper, L. L. Folgoc, M. J. Lee, M. Heinrich, K. Misawa, K. Mori, S. G. McDonagh, N. Hammerla, B. Kainz, B. Glocker, and D. Rueckert, “Attention u-net: Learning where to look for the pancreas,”ArXiv, vol. abs/1804.03999, 2018.
19. H. Zhang, I. Goodfellow, D. Metaxas, and A. Odena, “Self-attention generative adversarial networks,” in Proceedings of the 36th International Conference on Machine Learning(K. Chaudhuri and R. Salakhutdinov, eds.), vol. 97 of proceedings of Machine Learning Research, pp. 7354–7363, PMLR, 09–15 Jun 2019.
Figures
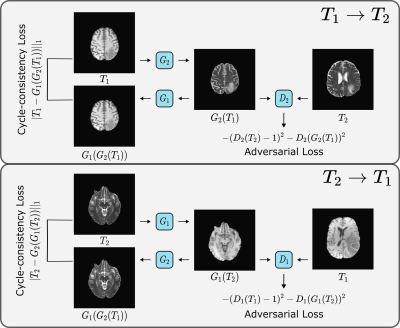
The unpaired translation method is based on two ResViT models with generators $$$G_1, G_2$$$ and two PatchGAN discriminators $$$D_1, D_2$$$. $$$G_2$$$ learns to translate a T1-weighted image to a T2-weighted image of the same cross-section that is indistinguishable from actual T2-weighted images from separate subjects, while $$$D_2$$$ learns to distinguish synthesized versus actual T2-weighted images. Likewise, $$$G_1$$$ learns to generate a T1-weighted image from a T2-weighted image, whereas $$$D_1$$$ learns to distinguish between synthetic and real T1-weighted images.