0551
Accelerated 2D Cardiac MRF Using a Self-Supervised Deep Image Prior Reconstruction1Radiology, University of Michigan, Ann Arbor, MI, United States, 2Biomedical Engineering, University of Michigan, Ann Arbor, MI, United States
Synopsis
A physics-based deep learning reconstruction is proposed for cardiac MRF that is based on the deep image prior framework, which employs self-supervised training and does not require additional in vivo training data. Calculation of the forward model is accelerated using a neural network to rapidly output MRF signal timecourses (in place of a Bloch simulation) and low-rank modeling to reduce the number of NUFFT operations. The proposed reconstruction enables cardiac T1/T2 mapping during a shortened 5-heartbeat breathhold and 150ms acquisition window.
Introduction
Several approaches have been proposed for Magnetic Resonance Fingerprinting (MRF)1 that use neural networks to shorten computation times and mitigate artifacts and noise2–5. One technique was recently applied to cardiac MRF6 using a fully-connected network to estimate T1 and T2 values from pixelwise timecourses7. However, convolutional networks that leverage both temporal and spatial information may achieve better performance, as shown for high-resolution MRF mapping in the brain4,8, but often require collection of additional in vivo scans for training that may be difficult to obtain. This work introduces a deep image prior (DIP) reconstruction for cardiac MRF that does not require prior training, which is used to shorten the breathhold and diastolic acquisition window for simultaneous 2D T1 and T2 mapping.Methods
The deep image prior (DIP) framework9 was modified using a U-net10 to generate cardiac MRF tissue property maps (Figure 1a). The input is a tensor of uniformly distributed random numbers between (-0.1, 0.1) that remains fixed during training, with size NxNxD, where N is the matrix size and D is the channel depth (this work sets D=10). The U-net performs 2D convolutions with five downsampling/upsampling paths and skip connections, which enforces a smoothness prior on the maps. The U-net outputs T1, T2, and proton density (M0) maps and is trained in a self-supervised manner (Figure 1b). A time series of images is obtained by inputting the maps into a fully-connected “fingerprint generator network (FGN),” which is pre-trained to rapidly output MRF timecourses for any T1, T2, and cardiac rhythm, as described previously11, with slice profile and preparation pulse efficiency corrections12,13. Next, the MRI encoding model is simulated, including multiplication by proton density scaling and coil sensitivity maps, spiral k-space undersampling, projection onto a low-rank temporal subspace14 (derived using the FGN) to reduce the number of NUFFT15 operations, and coil combination. This process yields aliased images in the temporal subspace that are compared to acquired images using a normalized l1-l2 loss to update the U-net. In all experiments, training was performed for 10,000 epochs with an Adam optimizer (learning rate 0.001) and 10% dropout, implemented using TensorFlow 2.6.0 with Keras on a GPU.The reconstruction was evaluated in simulations using the XCAT phantom16. Scan parameters were similar to previous work (FISP-MRF17, multiple inversions and T2 preparations, FA 4-25°, TR 5.3ms, 48-fold undersampled spiral with pseudo golden angle rotation, 192x192, 300x300mm2 FOV) with a 15-heartbeat breathhold and 254ms acquisition window18. Shortened scans were also simulated with a 5-heartbeat breathhold and 254ms, 200ms, 150ms, 100ms, and 50ms acquisition windows. Maps were reconstructed using dot product matching1, a low-rank reconstruction with locally low rank (6x6 patch size, λ1=0.02 relative to maximum intensity) and spatial total variation (λ2=0.002) regularization19, and the proposed DIP method, and nRMSE was calculated relative to ground truth maps. The T2 layer of the ISMRM/NIST MRI system phantom20 was scanned at 1.5T (Sola, Siemens Healthineers) using the original cardiac MRF sequence and a shortened scan (5HB breathhold, 150ms acquisition window). MRF T1 and T2 values were compared to inversion recovery and spin echo measurements. Six healthy subjects were also scanned at 1.5T with data acquired at a mid-ventricular slice using original and shortened MRF scans and conventional sequences (MOLLI and T2-prepared bSSFP)21,22, and T1 and T2 values were measured in the myocardial septum.
Results
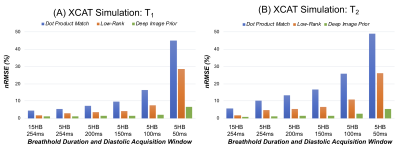
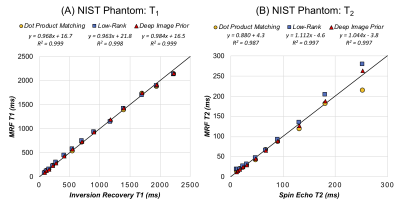
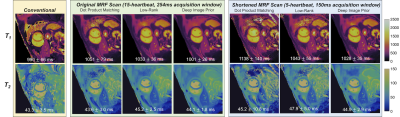
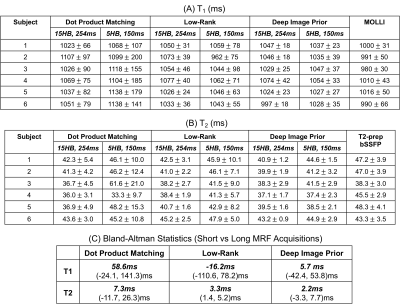
In simulations (Figure 2), scans with shorter breathholds and acquisition windows yielded higher nRMSE. DIP outperformed dot product matching and the low-rank reconstruction in all cases. Using a cutoff of nRMSE<2%, the shortest achievable scan was a 5-heartbeat breathhold and 150ms acquisition window, which was used for subsequent experiments. Phantom results are shown in Figure 3 using the shortened scan. The DIP reconstruction was most consistent with reference measurements, especially for T2, with the best-fit line having a slope closer to 1 and y-intercept closer to 0. Figure 4 shows representative maps using original and shortened MRF acquisitions. The DIP reconstruction reduced noise and aliasing artifacts with a visible improvement over comparison methods for the shortened scan. As shown in Table 1, DIP yielded lower standard deviations and a closer agreement between mean T1 and T2 values using original versus shortened MRF acquisitions, which is reflected in the Bland-Altman analysis in Table 1c.Discussion
A deep image prior reconstruction was introduced for cardiac MRF that enables a shortened 5-heartbeat breathhold and 150ms acquisition window for 2D T1/T2 mapping, which may be useful in patients who cannot perform long breathholds or have rapid heart rates. Because the method is self-supervised, there is no need to collect additional in vivo scans for training, which is time-consuming and may not generalize to different cardiac rhythms (which affects T1 and T2 quantification). This work also used strategies to accelerate the calculation of the forward model, including using a neural network to rapidly output cardiac MRF timecourses rather than using a Bloch simulation, and using low-rank modeling to reduce the number of NUFFT operations. Ongoing work will evaluate this technique in patients and post-contrast mapping.Conclusion
A deep image prior reconstruction is introduced for cardiac MRF that does not require additional training data and enables a shortened 5-heartbeat breathhold and 150ms acquisition window.Acknowledgements
NIH R01HL146021; NSF/CBET 1553441; Siemens HealthineersReferences
- Ma D, Gulani V, Seiberlich N, et al. Magnetic resonance fingerprinting. Nature. 2013;495(7440):187-192.
- Cohen O, Zhu B, Rosen MS. MR fingerprinting Deep RecOnstruction NEtwork (DRONE). Magn Reson Med. 2018;80(3):885-894.
- Yang M, Jiang Y, Ma D, Mehta BB, Griswold MA. Game of Learning Bloch Equation Simulations for MR Fingerprinting. In: Proc. 26th Annu. Meet. ISMRM. Paris, France; 2018:673.
- Fang Z, Chen Y, Liu M, et al. Deep Learning for Fast and Spatially-Constrained Tissue Quantification from Highly-Accelerated Data in Magnetic Resonance Fingerprinting. IEEE Trans Med Imaging. 2019;38(10):2364-2374.
- Cao P, Cui D, Vardhanabhuti V, Hui ES. Development of fast deep learning quantification for magnetic resonance fingerprinting in vivo. Magn Reson Imaging. 2020;70:81-90.
- Hamilton JI, Jiang Y, Chen Y, et al. MR fingerprinting for rapid quantification of myocardial T1, T2, and proton spin density. Magn Reson Med. 2017;77(4):1446-1458.
- Hamilton JI, Currey D, Rajagopalan S, Seiberlich N. Deep learning reconstruction for cardiac magnetic resonance fingerprinting T1 and T2 mapping. Magn Reson Med. 2021;85(4):2127-2135.
- Fang Z, Chen Y, Hung S, Zhang X, Lin W, Shen D. Submillimeter MR fingerprinting using deep learning–based tissue quantification. Magn Reson Med. 2020;84(2):579-591.
- Lempitsky V, Vedaldi A, Ulyanov D. Deep Image Prior. In: Proceedings of the IEEE Computer Society Conference on Computer Vision and Pattern Recognition. IEEE Computer Society; 2018:9446-9454.
- Ronneberger O, Fischer P, Brox T. U-net: Convolutional networks for biomedical image segmentation. International Conference on Medical Image Computing and Computer-Assisted Invervention. Vol 9351. Springer Verlag; 2015:234-241.
- Hamilton JI, Seiberlich N. Machine Learning for Rapid Magnetic Resonance Fingerprinting Tissue Property Quantification. Proc IEEE. 2020;108(1):69-85.
- Ma D, Coppo S, Chen Y, et al. Slice profile and B1 corrections in 2D magnetic resonance fingerprinting. Magn Reson Med. 2017;78(5):1781-1789.
- Hamilton JI, Jiang Y, Ma D, et al. Investigating and reducing the effects of confounding factors for robust T1 and T2 mapping with cardiac MR fingerprinting. Magn Reson Imaging. 2018;53:40-51.
- McGivney DF, Pierre E, Ma D, et al. SVD compression for magnetic resonance fingerprinting in the time domain. IEEE Trans Med Imaging. 2014;33(12):2311-2322.
- Fessler J, Sutton B. Nonuniform fast Fourier transforms using min-max interpolation. IEEE Trans Signal Process. 2003;51(2):560-574.
- Segars WP, Sturgeon G, Mendonca S, Grimes J, Tsui BMW. 4D XCAT phantom for multimodality imaging research. Med Phys. 2010;37(9):4902-4915.
- Jiang Y, Ma D, Seiberlich N, Gulani V, Griswold MA. MR fingerprinting using fast imaging with steady state precession (FISP) with spiral readout. Magn Reson Med. 2015;74(6):1621-1631.
- Hamilton JI, Pahwa S, Adedigba J, et al. Simultaneous Mapping of T1 and T2 Using Cardiac Magnetic Resonance Fingerprinting in a Cohort of Healthy Subjects at 1.5T. J Magn Reson Imaging. 2020;52(4):1044-1052.
- Lima da Cruz G, Bustin A, Jaubert O, Schneider T, Botnar RM, Prieto C. Sparsity and locally low rank regularization for MR fingerprinting. Magn Reson Med. 2019;81(6):3530-3543.
- Keenan KE, Stupic KF, Boss M, et al. Comparison of T1 measurement using ISMRM/NIST system phantom. In: Proceedings of the 24th Annual Meeting of ISMRM, Singapore; 2016:3290.
- Messroghli DR, Radjenovic A, Kozerke S, Higgins DM, Sivananthan MU, Ridgway JP. Modified Look-Locker inversion recovery (MOLLI) for high-resolution T1 mapping of the heart. Magn Reson Med. 2004;52(1):141-146.
- Giri S, Chung Y-C, Merchant A, et al. T2 quantification for improved detection of myocardial edema. J Cardiovasc Magn Reson. 2009;11(1):56.
Figures
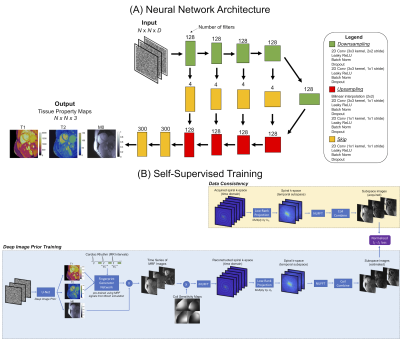
Figure 1: (a) A U-net that has 2D convolutional layers, 5 downsampling/upsampling paths, and skip connections is used to generate cardiac MRF T1, T2, and M0 maps. (b) The estimated maps, along with the subject's cardiac RR interval timings obtained from the ECG, are input to a pre-trained “fingerprint generator network," which outputs a time series of MRF images. These images are used to simulate the MRI encoding model, including coil sensitivities, spiral undersampling, and low-rank projection. A normalized l1-l2 loss is calculated in the image domain to update the U-net.