0545
VERDICT MRI estimates match histopathology features in brain tumours1Centre for Medical Image Computing, Computer Science Department, University College London, London, United Kingdom, 2Neuroradiology Unit and CERMAC, Vita-Salute San Raffaele University and IRCCS Ospedale San Raffaele, Milan, Italy, 3Department of Neurosurgery and Gamma Knife Radiosurgery, Vita-Salute San Raffaele University and IRCCS Ospedale San Raffaele, Milan, Italy, 4Pathology Unit, IRCCS Ospedale San Raffaele, Milan, Italy, 5Philips Healthcare, Milan, Italy, 6Cardiff University Brain Research Imaging Centre (CUBRIC), School of Psychology, Cardiff University, Cardiff, United Kingdom, 7School of Computer Science and Informatics, Cardiff University, Cardiff, United Kingdom, 8Clinical Imaging Sciences Centre, Brighton and Sussex Medical School, Brighton, United Kingdom
Synopsis
We recently adapted the VERDICT framework to characterize both the core and peritumoural areas of brain tumours. We report here its first clinical application in the differentiation of brain tumour histotypes. Comparing groups of lesions with increasing aggressiveness (from lower to higher grades to metastases) we observed a significant increase in the intracellular and vascular fraction in the lesion core. VERDICT maps matched the features showed by histopathology in lower grades and in metastases; in the most heterogeneous higher grades, VERDICT maps showed differences between subregions compatible with histopathology results in multiple biopsy samples.
Introduction
The characterization of brain tumours plays a fundamental role for diagnosis, treatment planning and assessment of therapy effects, but standard non-invasive imaging has limited specificity in differentiating tumour types and invasive histopathology techniques are still the gold standard.1 Vascular, Extracellular, and Restricted Diffusion for Cytometry in Tumors (VERDICT) is a framework for multi-compartment modeling of tumour tissues from diffusion MRI (dMRI).2 It has mainly been applied to body cancer3-5, but we have recently adapted it to study brain tumours, which is more challenging due to the higher microstructural complexity of brain tissues.6 Here we show the first clinical application of VERDICT in the differentiation of brain tumour histotypes, and compare VERDICT maps to histopathology in specific lesions and tumour subregions.Methods
We collected data from 21 patients with brain tumours, including one grade-1 subependymoma, five grade-2 astrocytomas (4 IDH wild-type, 1 IDH mutant), eight grade-3 astrocytomas (5 IDH wild-type, 3 IDH mutant), one grade-3 ependymoma, three grade-4 glioblastomas (IDH wild-type), two melanoma metastases and one radionecrosis (table 1).MRI was acquired at 3T (Ingenia CX, Philips Healthcare). The protocol included 3D-FLAIR images, post-contrast 3D T1-weighted images, and a series of dMRI scans including 10 shells with 3 diffusion directions each, b-values from 50 to 3500 s/mm2 and variable diffusion and echo times, followed by two shells with b = 711 and 3000 s/mm2 and 38 and 63 directions respecitvely.6 We outlined regions of interest (ROIs) using 3D-FLAIR images as a reference for the whole lesion and post-contrast 3D-T1 images for the contrast-enhancing core.
After MP-PCA denoising7, removal of Gibbs artifacts7, motion and distortion correction8, we used a two-stage approach. First, we fitted NODDI9 to the data in the last two shells to estimate the fraction of free diffusion fFW. Then, using the terminology in 10, we fitted the whole dMRI signal as:
$$E= f_{FW} e^{-b⋅ D_{FW} } + \big(1- f_{FW} \big) \big(f_{IC}⋅Sphere(R,D_{IC} )+f_{EES}⋅Zeppelin(D_ \| ,D_ \bot ,dir)+f_{VASC}⋅Astrosticks(D_{VASC} )\big)$$
where fIC, fEES and fVASC are the signal fractions of the intracellular, extracellular, and vascular compartments, DIC, D∥, D⊥ and DVASC the diffusivity within each of them (with the constrain DVASC > 9·10-9 m2/s), fFW is the fraction of free water, constrained to the value estimated by NODDI, and DFW = 3·10-9 m2/s.
The tumours were classified as metastases, higher grades (HG) and lower grades (LG) according to histopathological diagnosis, and the corresponding peritumoural areas as vasogenic oedemas, infiltrative oedemas, and LG infiltrative periphery respectively. We compared the VERDICT metrics among these groups using the Wilcoxon rank-sum test. We also visually compared VERDICT maps to histopathological images in two cases that underwent gross resection and in three cases with stereotactic biopsy.
Results
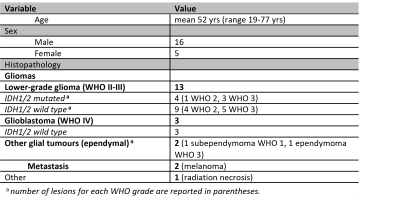
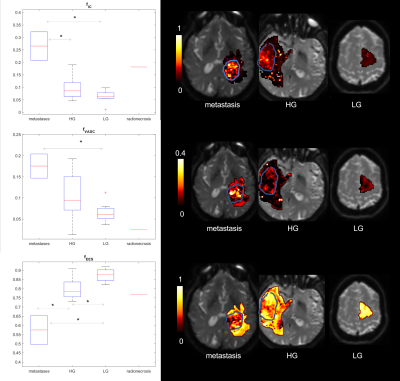
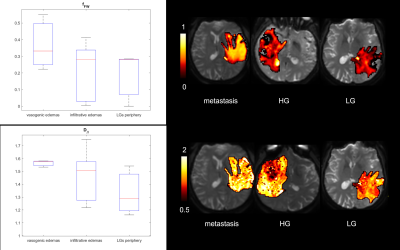
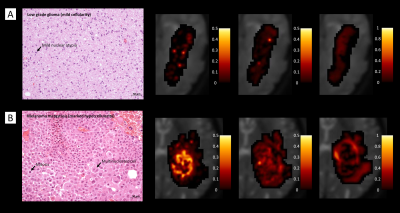
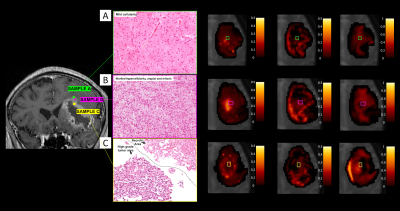
Figure 1 shows boxplots of the VERDICT signal fractions in the tumour core, along with representative maps from for the different histotypes. Metastases had the highest fIC and fVASC values, followed by HG and LG. Consequently, LG had the highest fEES, followed by HG and metastases. All the differences between metastases and LG were statistically significant, as well as the difference in fVASC and fEES between metastases and LG and in fEES between HG and LG. The radionecrosis case had very low fVASC and high fEES and fFW values.11-12 In the peritumoural areas, we observed a consistent trend towards higher fFW, D∥ and D⊥ values in vasogenic oedemas, followed by infiltrative oedemas and LG periphery (figure 2).VERDICT maps from a LG and a metastasis are shown in figure 3 along with histopathology images from gross resection. In the LG, where histopathology shows mild cellularity, fIC and fVASC are both very low. In the metastasis, where histopathology shows marked hypercellularity, there are areas of very high fIC and fVASC in the core of the tumour; fFW is high in some intra-tumoural spots that may correspond to necrosis and in the peritumoural area (vasogenic oedema). In the cases that underwent stereotactic biopsy, VERDICT maps reflected the trends shown by histopathology images across samples. An example is shown in figure 4. Sample B, with high grade features according to histopathology, had higher fIC than A and C, with low grade and heterogeneous appearance respectively. Sample C, where histopathology showed some necrosis, had the highest fFW.
Discussion and Conclusion
We presented the application of VERDICT to the differentiation between brain tumour types and subregions in comparison to histopathology. VERDICT provided significantly higher intracellular and vascular fractions in the most aggressive tumours, as expected. We also observed differences between peritumoural areas, with the trends that we expected, although not significant. VERDICT maps were compatible with the histological features in the most benign and most aggressive cases, and even in different subregions of the tumours when biopsy was performed in multiple samples. In future studies, we will optimise a shorter acquisition protocol13 and test the use of deep learning-based fitting14-15 to favour the clinical translation of the present approach. Larger validation studies with systematic point-to-point correlations should also be performed. These preliminary results hold promise for the non-invasive characterization of brain tumours by VERDICT, which would be an invaluable tool for diagnosis as well as for planning and evaluating treatments.Acknowledgements
EP is supported by EPSRC grant nr. EP/N021967/1
MP is supported by UKRI Future Leaders Fellowship (MR/T020296/1)
References
- Ellingson BM, Wen PY, van den Bent MJ, Cloughesy TF. Pros and cons of current brain tumor imaging. Neuro Oncol 2014;16(Suppl7):vii2-vii11
- Panagiotaki E, Walker-Samuel S, Siow B, et al. Noninvasive quantification of solid tumor microstructure using VERDICT MRI. Cancer Res 2014;74(7):1902-12
- Panagiotaki E, Chan RW, Dikaios N, et al. Microstructural characterization of normal and malignant human prostate tissue with vascular, extracellular, and restricted diffusion for cytometry in tumours magnetic resonance imaging. Invest Radiol. 2015;50(4):218-27
- Johnston EW, Bonet-Carne E, Ferizi U, et al. VERDICT MRI for Prostate Cancer: Intracellular Volume Fraction versus Apparent Diffusion Coefficient. Radiology. 2019;291(2):391-397
- Bailey C, Collins DJ, Tunariu N, et al. Microstructure Characterization of Bone Metastases from Prostate Cancer with Diffusion MRI: Preliminary Findings. Frontiers in Oncology. 2018;8(26).
- Figini M, Castellano A, Pieri V, et al. Estimation of the vascular fraction in brain tumors by VERDICT correlated with Perfusion MRI. Proc. Intl. Soc. Mag. Reson. Med. 2020; 28:4446
- Tournier JD, Smith R, Raffelt D, et al. MRtrix3: A fast, flexible and open software framework for medical image processing and visualisation. Neuroimage. 2019;202:116137
- Nilsson M, Szczepankiewicz F, Lampinen B, et al. An open-source framework for analysis of multidimensional diffusion MRI data implemented in MATLAB. Joint Annual Meeting ISMRM-ESMRMB, 2018. Paris, France. 5355.
- Zhang H, Schneider T, Wheeler-Kingshott CA, Alexander DC. NODDI: practical in vivo neurite orientation dispersion and density imaging of the human brain. Neuroimage. 2012;61(4):1000-16.
- Panagiotaki E, Schneider T, Siow B, et al. Compartment models of the diffusion MR signal in brain white matter: a taxonomy and comparison. Neuroimage. 2012;59(3):2241-54
- Castellano A, Anzalone N. Radiation and Chemotherapy Induced Injury. In: Barkhof F, Jager R, Thurnher M, Rovira Cañellas A, eds. Clinical Neuroradiology. Cham: Springer International Publishing; 2019: 1-29
- Perry A, Schmidt RE. Cancer therapy-associated CNS neuropathology: an update and review of the literature Acta Neuropathol. 2006;111:197–212
- Alexander DC. A general framework for experiment design in diffusion MRI and its application in measuring direct tissue-microstructure features. Magnetic Resonance in Medicine. 2008;60(2):439-48.
- de Almeida Martins JP, Nilsson M, Lampinen B, et al. Neural Networks for parameter estimation in microstructural MRI: a study with a high-dimensional diffusion-relaxation model of white matter microstructure. bioRxiv 2021.03.12.435163
- Gyori NG, Palombo M, Clark CA, Zhang H, Alexander DC. Training data distribution significantly impacts the estimation of tissue microstructure with machine learning. Magn Reson Med. 2021 Sep 21
Figures