0492
Inter-Rater Reliability of Fetal Lung Segmentation in High-Resolution Fetal Body Reconstructions1Center for MR Research, University Children's Hospital Zurich, Zurich, Switzerland, 2Neuroscience Center Zurich, University of Zurich, Zurich, Switzerland, 3Diagnostic Imaging and Intervention, University Children's Hospital Zurich, Zurich, Switzerland
Synopsis
Reliable measurements of the fetal lung volume are important for the prenatal assessment of various pathologies impacting fetal lung development. Here we compare the inter-rater reliability of fetal lung volume measurements segmented from native ssFSE scans to those from high-resolution fetal body reconstructions to determine if the high-resolution reconstructions provide a clinical benefit. We find that there is increased reliability in the volumes segmented from the high-resolution reconstructions, suggesting that high resolution reconstruction of the fetal lungs may enable more precise assessments of fetal lung volume.
Introduction
Reliable measurements of the fetal lung volume are important for the prenatal assessment of pathologies impacting the lungs such as congenital diaphragmatic hernia and pulmonary hypoplasia1,2. However, fetal lung segmentation is challenging due to the presence of fetal motion, and images acquired in non-standard planes. Recently it has become possible to create high-resolution (HR) reconstructions of the fetal body from multiple single-shot fast spin echo (ssFSE) scans. While preliminary studies show the possible benefits of using fetal body reconstructions for analysis of fetal lung volumes3, it is uncertain if these reconstructions can provide a more accurate and reliable fetal lung segmentation. Here, we present an analysis of fetal lung volumes segmented from both the low-resolution ssFSE and HR images by three observers in order to test inter-observer variability in both sets of fetal lung images. We hypothesized that the inter-observer variability would be significantly lower in the HR recons in comparison to the LR scans.Methods
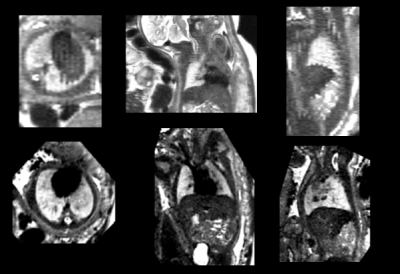
Clinical fetal MRI was acquired on 1.5T and 3T GE whole-body scanners using a T2-weighted ssFSE in 105 singleton pregnancies (19-37 gestational weeks) between September 2019 and July 2020. Scans in multiple orientations were acquired (axial, sagittal, coronal) with at least one scan in each orientation (0.5mmx0.5mmx3mm-5mm). The scans were reconstructed into HR volumes using a deformable slice-to-volume reconstruction technique to obtain images with a resolution of 0.5mmx0.5mmx0.5mm4. See Figure 1 for an example of an ssFSE scan and HR reconstruction. Three radiologists with varying experiences (between 1 and 10 years) with fetal MRI segmented the fetal lungs (left and right lung) in both the HR volume and in a self-chosen ssFSE scan, consistent with the standard clinical procedure. The radiologists rated the quality of each image, and only segmented the images they felt were of sufficient quality. In the HR fetal lungs, every 4th-5th slice was annotated, and the FSL and ITK’s morphological contour interpolation were used to create the final 3D segmentation5,6. In the ssFSE scan, every slice was segmented. The inter-observer reliability of the segmentations was then analyzed by looking at the difference in coefficients of variation (CV) between the three observers. A t-test was performed to determine if there was a difference in CV between the ssFSE and HR lung segmentations. Next, Bland-Altman plots were created to visualize the agreement between each of the three observers. Finally, a linear ANOVA model analyzing the CV was performed to look at the impact of gestational age (GA), magnet strength, and side of lung on the inter-observer reliability. All analysis was performed with R v4.0.3.Results
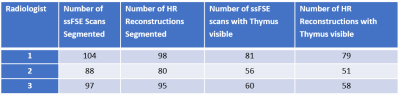
The number of scans segmented by each radiologist are shown in Table 1. There were 83 ssFSE cases and 80 HR cases where all radiologists provided a segmentation, including 67 cases where all radiologists provided both an ssFSE and HR segmentation.
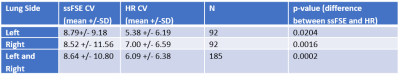
Interestingly, the inter-observer variability was similar in the left and right lungs in the ssFSE scans (p-value: p=0.65), but there was a difference in CV the left and right lungs in the HR scans, where the reliability was better for the left side in comparison to the right side (p=0.004, two-tailed paired t-test; see Table 2).
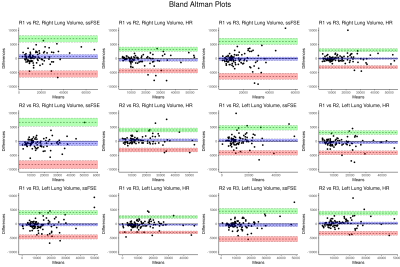
As expected, the inter-observer variability was lower in the HR recons in comparison to the ssFSE scans. When taking the left and right lung volume measurements as independent observations, the inter-observer variability of lung volume measurements was still significantly lower in the high resolution images. Figure 2 shows the Bland-Altman plots of each of the agreement between each observer. Visually, the difference between the raters is consistently smaller in the high-resolution images (dotted lines: 25th and 75th quartile, shaded regions: confidence interval).
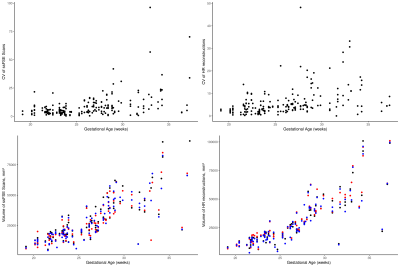
Finally, the linear ANOVA analysis revealed that in both the ssFSE and HR scans, gestational age explained much of the variance in segmented lung volume (ssFSE: p<0.001; HR: p<0.001), where the older fetuses were more challenging to segment. The lung laterality and MR field strength did not contribute significantly to inter-observer variance. See Figure 3 for a plot of volume vs GA and CV vs GA. Note that the fetuses included in this study contain both normal and pathological fetal lungs.
Discussion and Conclusion
While some of the HR reconstructions were not segmented due to low quality, the segmentations of the remaining HR reconstructions were more reliable, demonstrating more agreement between the three observers. The improvement in reliability from the HR reconstructions may arise from the motion correction of the images and orientation to a standard imaging plane, and may be particularly helpful for lung segmentation in older fetuses, in whom the variability was higher. In addition, the observers were free to choose from the several acquired ssFSE scans, further increasing the opportunity for variability. However, further work is needed to improve the image quality of the HR reconstructions which could not be segmented due to poor quality.In the future, an analysis of the normal lungs vs pathological lungs also needs to be completed. In conclusion, the HR reconstructions provide a more reliable segmentation of the fetal lungs, but do not increase the number of cases in which a manual segmentation is possible.
Acknowledgements
We would like to acknowledge the OPO Foundation, Anna Müller GrocholskiFoundation, the Foundation for Research in Science and the Humanities at the UZH, EMDO Foundation, HaslerFoundation, FZK Grant, and ZNZ PhD Grant for their support.References
1. Szpinda, M. et al. Volumetric growth of the lungs in human fetuses: an anatomical, hydrostatic and statistical study. Surg Radiol Anat 36, 813–820 (2014).
2. Debus, A. et al. Fetal Lung Volume in Congenital Diaphragmatic Hernia: Association of Prenatal MR Imaging Findings with Postnatal Chronic Lung Disease. Radiology 266, 887–895 (2013).
3. Davidson, J. et al. Motion corrected fetal body MRI provides reliable 3D lung volumes in normal and abnormal fetuses. https://www.authorea.com/users/425087/articles/538415-motion-corrected-fetal-body-mri-provides-reliable-3d-lung-volumes-in-normal-and-abnormal-fetuses?commit=a298b2952b75b2cf5fbc50496aaad73676148007 (2021) doi:10.22541/au.163251136.65436407/v1.
4. Uus, A. et al. Deformable Slice-to-Volume Registration for Motion Correction of Fetal Body and Placenta MRI. IEEE Trans Med Imaging 39, 2750–2759 (2020).
5. Jenkinson, M., Beckmann, C. F., Behrens, T. E. J., Woolrich, M. W. & Smith, S. M. FSL. Neuroimage 62, 782–790 (2012).
6. Pieper, S., Lorensen, B., Schroeder, W. & Kikinis, R. The NA-MIC Kit: ITK, VTK, Pipelines, Grids and 3D Slicer as An Open Platform for the Medical Image Computing Community. in 3rd IEEE International Symposium on Biomedical Imaging: Macro to Nano, 2006. 698–701 (IEEE, 2006). doi:10.1109/ISBI.2006.1625012.
Figures