0435
Rapid Quantitative Imaging Using Wave-Encoded Model-Based Deep Learning for Joint Reconstruction1Athinoula A. Martinos Center for Biomedical Imaging, Charlestown, MA, United States, 2Department of Radiology, Harvard Medical School, Boston, MA, United States, 3Fetal-Neonatal Neuroimaging & Developmental Science Center, Boston Children’s Hospital, Boston, MA, United States, 4National Nanotechnology Center, Pathum Thani, Thailand, 5Harvard/MIT Health Sciences and Technology, Cambridge, MA, United States
Synopsis
We propose a wave-encoded model-based deep learning (wave-MoDL) method for joint multi-contrast image reconstruction with volumetric encoding using an interleaved look-locker acquisition sequence with T2 preparation pulse (3D-QALAS). Wave-MoDL enables a 2-minute acquisition at R=4x3-fold acceleration using a 32-channel array to provide T1, T2, and proton density maps at 1 mm isotropic resolution, from which standard contrast-weighted images can also be synthesized.
Introduction
Wave-controlled aliasing in parallel imaging (wave-CAIPI)1,2 employs extra sinusoidal gradient modulations during the readout to harness coil sensitivity variations in all three dimensions and achieve higher accelerations. Recent wave-encoded model-based deep learning (wave-MoDL)3 has incorporated wave-CAIPI strategy into MoDL4,5 reconstruction that leverages convolutional neural networks (CNNs) and parallel imaging forward model to help denoise and unalias undersampled data. 3D-QuAntification using an interleaved Look-Locker Acquisition Sequence with T2 preparation pulse (3D-QALAS) enables high-resolution and simultaneous T1, T2, and proton density (PD) parameter mapping6–8. However, encoding limitations stemming from multi-contrast sampling at high resolution substantially lengthens 3D-QALAS acquisitions, e.g. 11 minutes for 1mm iso resolution at R=2-fold acceleration9. Combination of parallel imaging with compressed sensing has recently enabled a 6-minute scan9 at R=3.8. Unfortunately, pushing the acceleration further has not been possible due to g-factor penalty and intrinsic SNR limitations. In this abstract, we dramatically push the acceleration to R=12-fold to provide a 2-minute comprehensive quantitative exam by extending wave-MoDL to jointly reconstruct multi-contrast 3D-QALAS images, which reduces g-factor loss using wave-encoding and boosts SNR with MoDL. To use the complementary information across contrasts, we employ a CAIPI sampling pattern10 across multiple contrasts. From a 2-minute 3D-QALAS scan, wave-MoDL allows for estimation of high-quality T1, T2, and PD parameter maps, which can be used for synthesizing standard contrast-weighted images that can be used for clinical reads and subsequent analysis using existing softwares (e.g., FreeSurfer)11,12. Data/code can be found in: https://anonymous.4open.science/r/Wave-MoDL-Example-Code-ISMRM-2022-6DA0.Methods
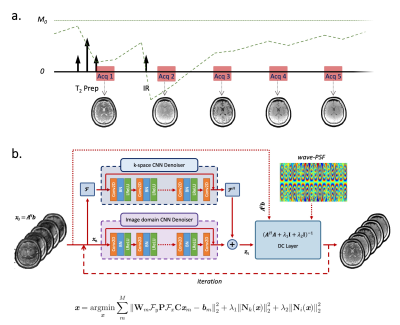
Sequence and Reconstruction. Figure 1a and Figure 1b show the 3D-QALAS sequence diagram and the proposed network architecture for wave-MoDL, respectively. We use unrolled CNNs to constrain the reconstruction in both the image- and k-spaces, which improves the performance of the entire network3,5,13,14. Wave-MoDL for multi-contrast image reconstruction can be described as follows.$$\textit{x}=\underset{\textit{x}}{\mathrm{argmin}}\sum_{m}^{M}{{\left\|\mathbf{W}_{m}\mathbf{\mathcal{F}}_y\mathbf{P}\mathbf{\mathcal{F}}_x\mathbf{C}\textit{x}_{m}-\textit{b}_{m}\right\|}_2^2}+\lambda_1{\left\|\mathit{\mathbf{N}}_k(\textit{x})\right\|}_2^2+\lambda_2{\left\|\mathit{\mathbf{N}}_i(\textit{x})\right\|}_2^2$$ $$=\underset{\textit{x}}{\mathrm{argmin}}\sum_{m}^{M}{{\left\|\mathbf{A}_{m}\textit{x}_{m}-\textit{b}_{m}\right\|}_2^2}+\lambda_1{\left\|\mathit{\mathbf{N}}_k(\textit{x})\right\|}_2^2+\lambda_2{\left\|\mathit{\mathbf{N}}_i(\textit{x})\right\|}_2^2$$
where $$$\textit{x}=\left\{\textit{x}_m|m=1...M\right\}$$$ are the reconstructed images, M is the number of contrasts, $$$\mathbf{W}_{m}$$$ is the subsampling mask for the m-th contrast, $$$\mathbf{P}$$$ is the wave point spread function in the kx-y-z hybrid domain, $$$\mathbf{C}$$$ is the coil sensitivity map, $$$\textit{x}_{m}$$$ is m-th contrast image, and $$$\textit{b}_{m}$$$ is the m-th subsampled data, $$$\mathit{\mathbf{N}}_k$$$ and $$$\mathit{\mathbf{N}}_i$$$ represent residual CNNs in the k-space and image-domain, respectively.
Data. The dataset includes 32-channel 3D-QALAS sagittal images acquired on a 3T Siemens Prisma scanner, consisting of five contrasts as shown in Figure 1a, for 10 subjects at 1mm-iso resolution. To reduce the chance of potential motion artifacts, data were acquired at R=2 to limit the scan time to 11minutes. High-SNR reference images were reconstructed by GRAPPA15 for five different contrast images separately. To train/validate/test the network, 8/1/1 subjects were used, respectively. The data were retrospectively subsampled to R=4x3, corresponding to 1:52-minute quantitative exam, where 5-cycle cosine and sine wave-encodings were applied in both ky and kz directions with 16.5mT/m of Gmax at 347Hz/pixel bandwidth. CAIPI sampling patterns were employed to exploit complementary k-space information across the multiple contrasts. To take into account the signal intensity differences between the contrasts, the contrast images were weighted by [3.26,2.36,1.57,1.12,1], calculated by the signal norm ratio of each contrast image in the training dataset. This allowed each contrast to contribute to the loss function in similar amounts. A CAIPI sampling pattern across the multi contrasts was applied to multi-contrast MoDL and multi-contrast wave-MoDL to use the complementary information from other contrasts, while a fixed sampling pattern was applied to five different contrasts for SENSE and wave-CAIPI since these algorithms reconstruct each image independently.
Results & Discussion
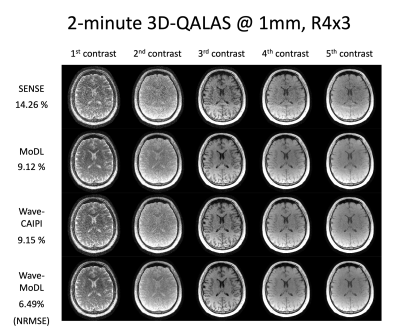
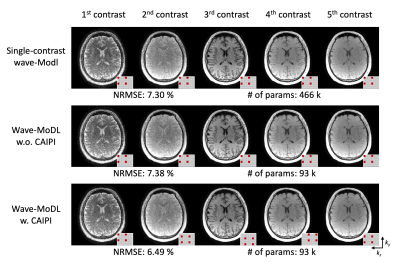
Figure 2 shows the multi-contrast image reconstruction using SENSE, MoDL, wave-CAIPI, and wave-MoDL at R=4x3-fold acceleration. SENSE suffers from aliasing artifacts and noise amplification, while MoDL and wave-CAIPI mitigated noise amplification and reduced the NRMSE significantly. Wave-MoDL further reduced NRMSE to 6.49%, which is a 1.4-fold improvement with respect to wave-CAIPI.Figure 3 shows the wave-MoDL results according to changing the sampling pattern. Multi-contrast wave-MoDL with CAIPI sampling pattern across multi-contrast shows the lowest NRMSE with 5-fold less number of network parameters with respect to single-contrast wave-MoDL.
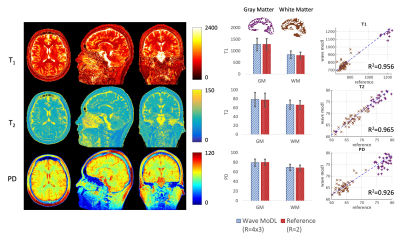
Figure 4 shows the T1, T2, and PD quantification results. To evaluate the accuracy, we calculated averaged values and standard deviation in white matter (WM, brown) and gray matter (GM, purple) segmented by FreeSurfer11,12. In the last column, we selected 50 boxes that have 5x5x5 size in WM and GM and plotted the quantified values using the wave-MoDL results over the reference R=2 acquisition. The plotted graphs demonstrate that the estimated T1, T2, and PD values are well aligned with the reference.
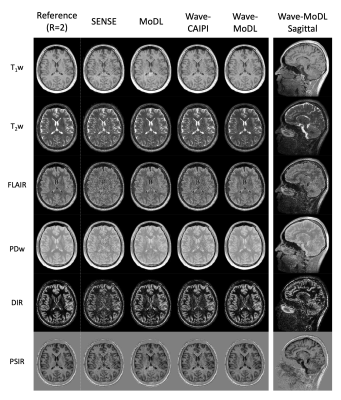
Figure 5 shows the synthesized T1w, T2w, FLAIR, PDw, double inversion recovery (DIR), and phase-sensitive inversion recovery (PSIR) images using the quantified T1, T2, and PD maps. The results demonstrate that wave-MoDL can mitigate the noise amplification and provide synthetic images that match well to the reference, compared with SENSE, MoDL, and wave-CAIPI.
Conclusion
We introduced joint multi-contrast image reconstruction using wave-MoDL for 3D-QALAS. Joint multi-contrast wave-MoDL markedly improved image quality, thus enabling a high acceleration of R=12 with a 32-channel coil array. Wave-MoDL for joint multi-contrast image reconstruction enabled a 2-minute 3D-QALAS acquisition at 1mm-iso resolution and permitted simultaneous T1, T2, and PD quantification as well as synthesis of standard contrast-weighted images with high fidelity.Acknowledgements
This work was supported by research grants NIH R01 EB028797, R03 EB031175, U01 EB025162, P41 EB030006, U01 EB026996, and the NVidia Corporation for computing support.References
1. Bilgic B, Gagoski BA, Cauley SF, et al. Wave-CAIPI for highly accelerated 3D imaging. Magn. Reson. Med. 2015;73:2152–2162 doi: 10.1002/mrm.25347.
2. Gagoski BA, Bilgic B, Eichner C, et al. RARE/turbo spin echo imaging with simultaneous multislice Wave-CAIPI: RARE/TSE with SMS Wave-CAIPI. Magn. Reson. Med. 2015;73:929–938 doi: 10.1002/mrm.25615.
3. Cho J, Tian Q, Frost R, Chatnuntawech I, Bilgic B. Wave-Encoded Model-Based Deep Learning with Joint Reconstruction and Segmentation. In: Proceedings of the 29th Scientific Meeting of ISMRM. Online Conference; 2021. p. 1982.
4. Aggarwal HK, Mani MP, Jacob M. MoDL: Model Based Deep Learning Architecture for Inverse Problems. 2017 doi: 10.1109/TMI.2018.2865356.
5. Aggarwal HK, Mani MP, Jacob M. MoDL-MUSSELS: Model-Based Deep Learning for Multi-Shot Sensitivity Encoded Diffusion MRI. IEEE Trans. Med. Imaging 2020;39:1268–1277 doi: 10.1109/TMI.2019.2946501.
6. Kvernby S, Warntjes MJB, Haraldsson H, Carlhäll C-J, Engvall J, Ebbers T. Simultaneous three-dimensional myocardial T1 and T2 mapping in one breath hold with 3D-QALAS. J. Cardiovasc. Magn. Reson. 2014;16:102 doi: 10.1186/s12968-014-0102-0.
7. Kvernby S, Warntjes M, Engvall J, Carlhäll C-J, Ebbers T. Clinical feasibility of 3D-QALAS – Single breath-hold 3D myocardial T1- and T2-mapping. Magn. Reson. Imaging 2017;38:13–20 doi: 10.1016/j.mri.2016.12.014.
8. Fujita S, Hagiwara A, Hori M, et al. Three-dimensional high-resolution simultaneous quantitative mapping of the whole brain with 3D-QALAS: An accuracy and repeatability study. Magn. Reson. Imaging 2019;63:235–243 doi: 10.1016/j.mri.2019.08.031.
9. Fujita S, Hagiwara A, Takei N, et al. Accelerated Isotropic Multiparametric Imaging by High Spatial Resolution 3D-QALAS With Compressed Sensing: A Phantom, Volunteer, and Patient Study. Invest. Radiol. 2021;56:292–300 doi: 10.1097/RLI.0000000000000744.
10. Breuer FA, Blaimer M, Mueller MF, et al. Controlled aliasing in volumetric parallel imaging (2D CAIPIRINHA). Magn. Reson. Med. 2006;55:549–556 doi: 10.1002/mrm.20787.
11. Dale AM, Fischl B, Sereno MI. Cortical Surface-Based Analysis: I. Segmentation and Surface Reconstruction. NeuroImage 1999;9:179–194 doi: 10.1006/nimg.1998.0395.
12. Desikan RS, Ségonne F, Fischl B, et al. An automated labeling system for subdividing the human cerebral cortex on MRI scans into gyral based regions of interest. Neuroimage 2006;31:968–980.
13. Eo T, Jun Y, Kim T, Jang J, Lee H-J, Hwang D. KIKI-net: cross-domain convolutional neural networks for reconstructing undersampled magnetic resonance images. Magn. Reson. Med. 2018;80:2188–2201 doi: 10.1002/mrm.27201.
14. Han Y, Sunwoo L, Ye JC. $k$ -Space Deep Learning for Accelerated MRI. IEEE Trans. Med. Imaging 2020;39:377–386 doi: 10.1109/TMI.2019.2927101.
15. Griswold MA, Jakob PM, Heidemann RM, et al. Generalized autocalibrating partially parallel acquisitions (GRAPPA). Magn. Reson. Med. 2002;47:1202–1210 doi: 10.1002/mrm.10171.
Figures