0394
Deep-learning-informed parameter estimation improves reliability of spinal cord diffusion MRI1Centre for Medical Image Computing, Department of Computer Science, University College London, London, United Kingdom, 2Radiomics Group, Vall d’Hebron Institute of Oncology, Vall d’Hebron Barcelona Hospital Campus, Barcelona, Spain, 3Queen Square MS Centre, Queen Square Institute of Neurology, Faculty of Brain Sciences, University College London, London, United Kingdom, 4NMR Research Unit, Queen Square Multiple Sclerosis Centre, Department of Neuroinflammation, UCL Queen Square Institute of Neurology, University College London, London, United Kingdom, 5Brain Connectivity Centre, IRCCS Mondino Foundation, Pavia, Italy, 6Department of Brain and Behavioural Sciences, University of Pavia, Pavia, Italy
Synopsis
The very low SNR in spinal cord diffusion MRI can make robust microstructure parameter estimation challenging. To address unreliable parameter estimations under such low SNR, we propose a deep-learning-informed maximum likelihood estimation (MLE) approach, where a deep-learning model is trained to initialise MLE efficiently and optimally. In testing NODDI-derived parameters, simulation and in vivo experiments suggest the DL-informed method can reduce outlier estimates from conventional grid-search MLE, and at the same time, avoid biases from pure DL estimation under the low SNR. The proposed method also speeds up the computation, making it a promising tool for future applications.
Introduction
Robust parameter estimation in microstructure imaging can be challenging when signal to noise ratio (SNR) is very low, as in spinal cord diffusion MRI. To estimate microstructure parameters, the conventional approach is maximum-likelihood estimation (MLE) based on non-linear optimisation1. However, the accuracy of this approach can depend highly on initialisation. With unfavourable initialisation, the optimisation could be stuck at suboptimal or local minima, resulting in outlier estimates that will affect following analysis. Several strategies have been developed to address this issue, such as conducting a grid search in the parameter space to find better initial guesses, or repeating the optimisation multiple times with different starting points. These methods, however, will increase the computation time substantially.Recently, deep-learning (DL) based methods have been developed as an alternative to the conventional MLE approach2,3. This method is known for its speed. A trained DL model can estimate microstructure parameters in large datasets almost instantly. However, a recent study suggests DL estimation can generate systematic bias in estimated parameters especially when SNR is low4.
To address the challenge of parameter estimation under low SNR, we propose a DL-informed MLE approach that combines the conventional MLE and the DL approaches synergistically. In the proposed approach, an artificial neural network (ANN) model is trained to initialise MLE efficiently and optimally, with the final solution determined by MLE, avoiding biases typically affecting pure DL estimation.
Methods
We evaluate the DL-informed MLE method for NODDI5 model estimation in the spinal cord. To map the noisy diffusion signals to NODDI-derived parameters, a fully connected ANN containing an input layer, 3 hidden layers (150 nodes for each layer), and an output layer with 3 outputs is designed to get the initial estimation of intra-neurite fraction ($$$f_{in}$$$), free water fraction ($$$f_{iso}$$$) and orientation dispersion index (odi). The following MLE optimisation accounts for a Rician noise model which is essential for low SNR data. To simplify optimisation, the fibre orientation is fixed to the principal eigenvector of diffusion tensor, given that the fibre in the spinal cord is highly aligned.The ANN is trained with simulated dataset generated from uniform coverage of $$$f_{in}$$$ and odi from 0 to 1, which have been suggested to be able to achieve lower estimation bias4; the distribution for $$$f_{iso}$$$ contains more samples below 0.4 as including high $$$f_{iso}$$$ samples for training will bias other parameters. Rician noise is added to the simulation to achieve an SNR of 10 for b=0 signal typically seen in spinal cord data6. The training sample size is 1 million.
Experiments
The DL-informed MLE is evaluated on both simulation and in vivo datasets, and compared to grid-search MLE (GS-MLE) in terms of computation time and estimation accuracy. In a conventional GS-MLE, the initialisation is decided by estimating the likelihoods of measurements given 125 uniform grid locations in the parameter space, and choosing the one that gives the highest likelihood.
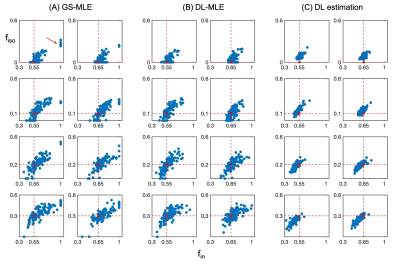
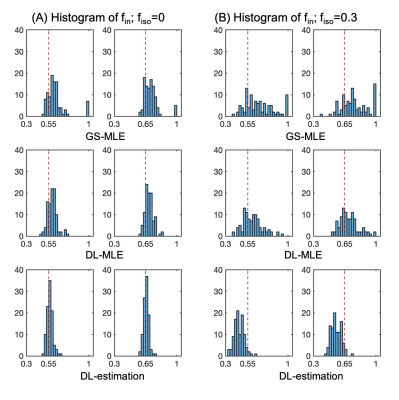
In vivo spinal cord data from a previous study were retrospectively analysed. These consisted of scans acquired with a 3T Philips Achieva scanner from 5 healthy subjects. A standard NODDI protocol is used for data acquisition with b = 0 (6 repetitions), b=711 (30 directions) and b=2855 (60 directions) s/mm2. Details about acquisition parameters and pre-processing steps (motion correction, segmentation) can be found here7. For simulated data, typical sets of tissue parameters are explored, with $$$f_{in}$$$ = [0.55, 0.65], $$$f_{iso} = [0, 0.1, 0.2, 0.3]$$$, and odi = 0.02 given in previous study7. For each set of tissue parameters, 100 random Rician noise realisations are generated to assess the accuracy and precision with SNR level of 10 for b=0 signal.
Results
The training time for the ANN model is about 7 mins on a single CPU; this trained model is applied to all new datasets to get initialisations for MLE which is done almost instantly. Without the time for grid search process voxel by voxel, the computation time for the DL-MLE method is about 1.75 times faster than the GS-MLE (Table.1).Results from simulation suggest that the DL-MLE method largely eliminates the outlier estimates of $$$f_{in}$$$ in GS-MLE, improving the precision of estimation especially when there is high CSF contamination; the direct DL-estimation, while improves the precision of estimates, generates a systematic bias in the estimation. Specifically, $$$f_{in}$$$ is underestimated when there is CSF contamination. (Fig.1-2)
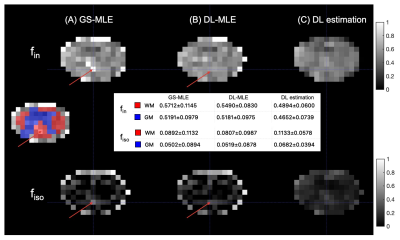
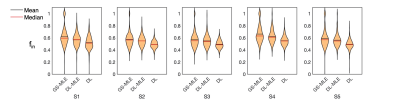
The in vivo results agree well with simulation findings. the DL-MLE gives an overall similar estimation to GS-MLE in the gray matter but is able to eliminate most of the outlier estimates in the WM, likely affected by CSF. The DL estimated $$$f_{in}$$$ are systematically lower than GS-MLE in all subjects. (Fig.3-4)
Conclusion
The DL-MLE method can improve the reliability of parameter estimations in the spinal cord where SNR is very low. While the method is developed for spinal cord imaging, it could also be extended to brain studies when data is acquired with high resolution hence lower SNR. Finally, the shorter computation time makes it a practical tool for future applications.Acknowledgements
The acquisition of the data used in this study was supported by the UCL Grand Challenges scheme, an initiative of the UCL School of Life and Medical Sciences, UCLH/UCL Biomedical Research Centre and the Specialist Biomedical Research Centres at Moorfields/UCL and Great Ormond Street/UCL, as well as by a programme grant from the UK MS Society (grant 892/08). The MRI scanner of the NMR Research Unit, Queen Square Multiple Sclerosis Centre, is supported by the National Institute for Health Research University College London Hospitals Biomedical Research Centre. CGWK receives grant funding from The Multiple Sclerosis Society (grant #77), Wings for Life (#169111), BRC (#BRC704/CAP/CGW), MRC (#MR/S026088/1), Ataxia UK. CGWK is a shareholder in Queen Square Analytics Ltd.
FG is supported by the investigator-initiated PREdICT study at the Vall d'Hebron Institute of Oncology (Barcelona), funded by AstraZeneca. AstraZeneca did not influence data acquisition and analysis, result interpretation and the decision to submit this work in its present form.
References
1. Alexander, D. C., Dyrby, T. B., Nilsson, M. & Zhang, H. Imaging brain microstructure with diffusion MRI: practicality and applications. NMR in Biomedicine vol. 32 186–198 (2019).
2. Golkov, V. et al. q-Space Deep Learning: Twelve-Fold Shorter and Model-Free Diffusion MRI Scans. IEEE Trans. Med. Imaging 35, 1344–1351 (2016).
3. Lin, Z. et al. Fast learning of fiber orientation distribution function for MR tractography using convolutional neural network. Med. Phys. 46, 3101–3116 (2019).
4. Gyori, N. G., Palombo, M., Clark, C. A., Zhang, H. & Alexander, D. C. Training data distribution significantly impacts the estimation of tissue microstructure with machine learning. Magn. Reson. Med. (2021) doi:10.1002/mrm.29014.
5. Zhang, H., Schneider, T., Wheeler-Kingshott, C. A. & Alexander, D. C. NODDI: Practical in vivo neurite orientation dispersion and density imaging of the human brain. Neuroimage 61, 1000–1016 (2012).
6. Grussu, F. et al. Relevance of time-dependence for clinically viable diffusion imaging of the spinal cord. Magn. Reson. Med. (2019) doi:10.1002/mrm.27463.
7. Grussu, F., Schneider, T., Zhang, H., Alexander, D. C. & Wheeler-Kingshott, C. A. M. Neurite orientation dispersion and density imaging of the healthy cervical spinal cord in vivo. Neuroimage (2015) doi:10.1016/j.neuroimage.2015.01.045.
Figures