0355
Self-supervised Training for Single-Shot Tumor Tracking in the Presence of Respiratory Motion1Medical Image And Data Analysis (MIDAS.lab), Department of Interventional and Diagnostic Radiology, University Hospital of Tuebingen, Tuebingen, Germany, 2University of Tuebingen, Tuebingen, Germany, 3Max Planck Institute for Intelligent Systems, Tuebingen, Germany, 4Department of Computer Science, Institute for Visual Computing, University of Tuebingen, Tuebingen, Germany
Synopsis
Real-time tumor tracking is a task of growing importance due to the increasing availability of modern linear accelerators paired with MR imaging, called MR-LINAC. Physiological motion can thereby impair focal treatment of moving lesions.Classical tracking approaches often work inadequately because they operate only at the pixel level and thus do not include image-level information.Contrarily, learning based tracking systems typically require a large, fully-annotated dataset which is an arduous task to create. In this work, we propose a framework for example-based single-shot tumor tracking, which is trained without presence of labels and investigated for lesion tracking under respiratory motion.
Introduction
Respiratory motion in the body trunk induces uncertainties in the location of a tumor during radiotherapy treatment. As a result, the efficacy of radiotherapy is reduced and higher radiation exposure with increased toxicity for the tumor surrounding tissues is attained. The MR-LINAC was introduced as a combination of an MR scanner and linear accelerator to enable MR-guided tracking of tumors during radiotherapy.Respiratory motion can induce cranio-caudal diaphragm displacements of up to 75 mm for deep inspiration1-3 with a concomitant non-rigid deformation of adjacent organs such as the lung and liver. Real-time tracking of these moving tumors with high spatial and temporal precision can therefore be challenging, but desired for delivery of localized beams to the target lesion.
Several strategies have been proposed to resolve lesion motion in MR images based on retrospective gating4-6, fusion of multi-orientation imaging7,8 or low-rank and model-based reconstructions9-12. Tumor tracking in these motion-resolved imaging data has grown to a large area of interest13-15. However, previously proposed tracking methods either rely on full supervision16, identification of markers17 or tracking via optical flow18 and non-rigid registrations19,20. Extensive tracking planning, setup and supervision is time- and cost-intensive and prone to errors. Therefore, tracking based on marking a single example image is desired.
More recently, machine learning based solutions were proposed to address the problem of full supervision by self-supervised learning of pixel-wise embeddings in anatomical images21 or unsupervised learning of deformation fields22. However, these approaches are computationally demanding during inference and do not meet the low latency requirement of $$$<$$$200 ms for real-time tumor tracking.
In this study we propose a self-supervised deep learning framework for real time landmark detection and tumor tracking that is able to accurately predict the position of a tumor based on a single initial example.
Methods
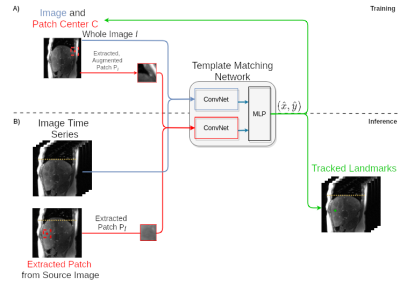
In this work, we investigate a real-time tumor tracking framework for respiratory motion-resolved MR-LINAC data which acts on a single labelled example. To allow label-free training (i.e. no supervision), we train a convolutional network on a within-image template matching task using self-supervision and targeted data augmentation.The template matching task consists of estimating the center position of image patches within source images.In detail, squared image patches $$$\mathbf{P_I}$$$ of predefined size are uniformly drawn from the respective source images $$$\mathbf{I}$$$ at a single time point.Both, patch at position $$$(x,y)$$$ and source image are then fed to a neural network to output an estimate of the patch center coordinates $$$\hat{C}=(\hat{x},\hat{y})$$$.Formally, we model this problem as the task to learn the conditional distribution of the center coordinates given source image and extracted patch under assumed Gaussian noise with mean $$${\boldsymbol{\mu}}$$$ and variance $$$\mathbf {\Sigma}$$$
$$\mathbf{P(C|P_I, I)}={\mathcal{N}}_{2}({\boldsymbol{\mu}},\mathbf{\Sigma}).$$
Self-supervised training can thus be conducted with the negative log likelihood sampling loss for location $$$x$$$ and $$$y$$$, respectively.
The proposed tracking network architecture (Fig. 1A) consists of two separate ConvNet encoders. Those encoders are based on the VGG16 architecture23 and pretrained on ImageNET24. The stacked encoder outputs are then fed into an MLP (3 layers) to predict the positional outputs $$$(\hat{x},\hat{y})$$$ and variance $$$(\hat{\sigma_x},\hat{\sigma_y})$$$.
Beyond identifying similar patches within the same image, our goal was to achieve stable prediction with generalization to cross-image tracking. Under the assumption that all training images contain similar objects, we hypothesize that this generalization can be achieved by regularization through data augmentation.Thus, we apply domain-specific data augmentation to the extracted image patches (Rotation: -5$$$^\circ$$$ to 5$$$^\circ$$$, affine scaling: 0.8 to 1.2, gamma contrast variation25: (0.7, 1.7)).
Tumor tracking can then be performed by feeding the time-resolved image sequence together with an initially selected example patch containing the lesion to track to the architecture (Fig. 1B).
The model is trained and evaluated on 2D sagittal motion resolved images acquired with spoiled GRE (TE/TR=1.8ms/3.6ms; flip angle=15°; bandwidth=670Hz/pixel; resolution=2$$$\times$$$2mm2; acquisition time/image=0.4s)26.
36 patients (60$$$\pm$$$9 years, 20 female) were scanned on a 3T MRI. A subject-level split of 60-20-20 (train-val-test) was used to minimize the negative log likelihood with the ADAM optimizer27 and an initial learning rate of 10-4 for 1000 epochs with patches of size (50$$$\times$$$50).
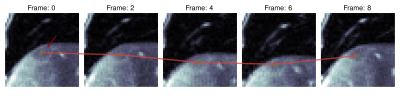
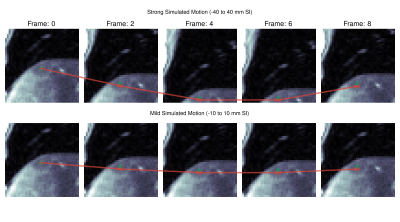
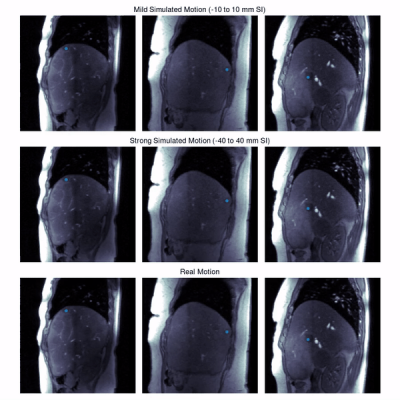
Experiments: We evaluate the tracking performance on 7 different lesions on both, simulated and real motion. Shallow (-10 to 10 mm) and deep (-40 to 40 mm) respiratory motion is simulated. For evaluation, lesions were manually annotated in all slices and frames by an experienced radiologist.
Results and Discussion
Quantitative evaluation is visualized in Fig. 2. A tracking accuracy of close to one pixel (for SI displacement even subpixel) is achieved. Performance is consistent between mild and strong simulated motion, with good agreement to realistic motion. Results of qualitative evaluations in different patients under simulated and realistic motion are shown in Fig. 3-5. They reveal sufficient tracking performance with runtime $$$\leq$$$ 5ms/frame, thus allowing real time tumor tracking.We acknowledge several limitations. In this work, we only focused on 2Dt data in a retrospective setting. In future works, we plan to extend the proposed approach for 3Dt tumor tracking with implementation on the MR-LINAC.
Conclusion
We proposed and evaluated a self-supervised training for example-based landmark tracking which does not require paired training samples. The proposed approach allows real-time tumor tracking in motion-resolved MRI.Acknowledgements
This project was partially funded by the Deutsche Forschungsgemeinschaft (DFG, German Research Foundation), grant number 438106095 and conducted under Germany’s Excellence Strategy – EXC-Number 2064/1 – Project number 390727645 and EXC-Number 2180 – Project number 390900677.References
1. Takazakura R, Takahashi M, Nitta N,
Murata K. Diaphragmatic motion in the sitting and supine positions: healthy
subject study using a vertically open magnetic resonance system. Journal of
Magnetic Resonance Imaging 2004;19(5):605-609.
2. Clifford MA, Banovac F, Levy E,
Cleary K. Assessment of hepatic motion secondary to respiration for computer
assisted interventions. Computer Aided Surgery 2002;7(5):291-299.
3. Catana C. Motion correction options
in PET/MRI. 2015 2015. p 212-223.
4. Breuer K, Meyer CB, Breuer FA,
Richter A, Exner F, Weng AM, Ströhle S, Polat B, Jakob PM, Sauer OA, others.
Stable and efficient retrospective 4D-MRI using non-uniformly distributed
quasi-random numbers. Physics in Medicine & Biology
2018;63(7):075002-075002.
5. Deng Z, Pang J, Yang W, Yue Y, Sharif
B, Tuli R, Li D, Fraass B, Fan Z. Four-dimensional MRI using three-dimensional
radial sampling with respiratory self-gating to characterize temporal
phase-resolved respiratory motion in the abdomen. Magnetic resonance in medicine
2016;75(4):1574-1585.
6. Paganelli C, Lee D, Kipritidis J,
Whelan B, Greer PB, Baroni G, Riboldi M, Keall P. Feasibility study on 3D image
reconstruction from 2D orthogonal cine-MRI for MRI-guided radiotherapy. Journal
of medical imaging and radiation oncology 2018;62(3):389-400.
7. Bjerre T, Crijns S, af Rosenschöld
PM, Aznar M, Specht L, Larsen R, Keall P. Three-dimensional MRI-linac
intra-fraction guidance using multiple orthogonal cine-MRI planes. Physics in
Medicine & Biology 2013;58(14):4943-4943.
8. Tryggestad E, Flammang A, Hales R,
Herman J, Lee J, McNutt T, Roland T, Shea SM, Wong J. 4D tumor centroid
tracking using orthogonal 2D dynamic MRI: implications for radiotherapy
planning. Medical physics 2013;40(9):091712-091712.
9. King AP, Buerger C, Tsoumpas C,
Marsden PK, Schaeffter T. Thoracic respiratory motion estimation from MRI using
a statistical model and a 2-D image navigator. Medical image analysis
2012;16(1):252-264.
10. Stemkens B, Tijssen RHN, De Senneville
BD, Lagendijk JJW, Van Den Berg CAT. Image-driven, model-based 3D abdominal
motion estimation for MR-guided radiotherapy. Physics in Medicine & Biology
2016;61(14):5335-5335.
11. Ong F, Zhu X, Cheng JY, Johnson KM,
Larson PEZ, Vasanawala SS, Lustig M. Extreme MRI: Large-scale volumetric dynamic
imaging from continuous non-gated acquisitions. Magnetic resonance in medicine
2020;84(4):1763-1780.
12. Huttinga NRF, Bruijnen T, van den Berg
CAT, Sbrizzi A. Nonrigid 3D motion estimation at high temporal resolution from
prospectively undersampled k-space data using low-rank MR-MOTUS. Magnetic
Resonance in Medicine 2021;85(4):2309-2326.
13. Hunt A, Hansen VN, Oelfke U, Nill S,
Hafeez S. Adaptive radiotherapy enabled by MRI guidance. Clinical Oncology
2018;30(11):711-719.
14. Cervino LI, Du J, Jiang SB. MRI-guided
tumor tracking in lung cancer radiotherapy. Physics in Medicine & Biology
2011;56(13):3773-3773.
15. Sakata Y, Hirai R, Kobuna K, Tanizawa
A, Mori S. A machine learning-based real-time tumor tracking system for
fluoroscopic gating of lung radiotherapy. Physics in Medicine & Biology
2020;65(8):085014-085014.
16. Tahmasebi N, Boulanger P, Noga M,
Punithakumar K. A fully convolutional deep neural network for lung tumor
boundary tracking in MRI. 2018 2018. p 5906-5909.
17. Booth J, Caillet V, Briggs A,
Hardcastle N, Angelis G, Jayamanne D, Shepherd M, Podreka A, Szymura K, Nguyen
DT, others. MLC tracking for lung SABR is feasible, efficient and delivers
high-precision target dose and lower normal tissue dose. Radiotherapy and
Oncology 2021;155:131-137.
18. Zachiu C, Papadakis N, Ries M, Moonen
C, de Senneville BD. An improved optical flow tracking technique for real-time
MR-guided beam therapies in moving organs. Physics in Medicine & Biology
2015;60(23):9003-9003.
19. Tahmasebi N, Boulanger P, Yun J,
Fallone G, Noga M, Punithakumar K. Real-Time Lung Tumor Tracking Using a CUDA
Enabled Nonrigid Registration Algorithm for MRI. IEEE journal of translational
engineering in health and medicine 2020;8:1-8.
20. Huttinga NRF, van den Berg CAT, Luijten
PR, Sbrizzi A. MR-MOTUS: model-based non-rigid motion estimation for MR-guided
radiotherapy using a reference image and minimal k-space data. Physics in
Medicine & Biology 2020;65(1):015004-015004.
21. Yan K, Cai J, Jin D, Miao S, Harrison
AP, Guo D, Tang Y, Xiao J, Lu J, Lu L. Self-supervised learning of pixel-wise
anatomical embeddings in radiological images. arXiv preprint arXiv:201202383
2020.
22. Lei Y, Fu Y, Wang T, Liu Y, Patel P,
Curran WJ, Liu T, Yang X. 4D-CT deformable image registration using multiscale
unsupervised deep learning. Physics in Medicine & Biology
2020;65(8):085003-085003.
23. Simonyan K, Zisserman A. Very Deep
Convolutional Networks for Large-Scale Image Recognition. 2015.
24. Russakovsky O, Deng J, Su H, Krause J,
Satheesh S, Ma S, Huang Z, Karpathy A, Khosla A, Bernstein M, others. Imagenet
large scale visual recognition challenge. International journal of computer
vision 2015;115(3):211-252.
25. Jung AB, Wada K, Crall J, Tanaka S,
Graving J, Reinders C, Yadav S, Banerjee J, Vecsei G, Kraft A, Rui Z, Borovec
J, Vallentin C, Zhydenko S, Pfeiffer K, Cook B, Fernández I, De Rainville F-M,
Weng C-H, Ayala-Acevedo A, Meudec R, Laporte M, others. imgaug. 2020.
26. Würslin C, Schmidt H, Martirosian P,
Brendle C, Boss A, Schwenzer NF, Stegger L. Respiratory motion correction in
oncologic PET using T1-weighted MR imaging on a simultaneous whole-body PET/MR
system. Journal of nuclear medicine 2013;54(3):464-471.
27. Kingma DP, Ba J.
Adam: A Method for Stochastic Optimization. 2017.
Figures